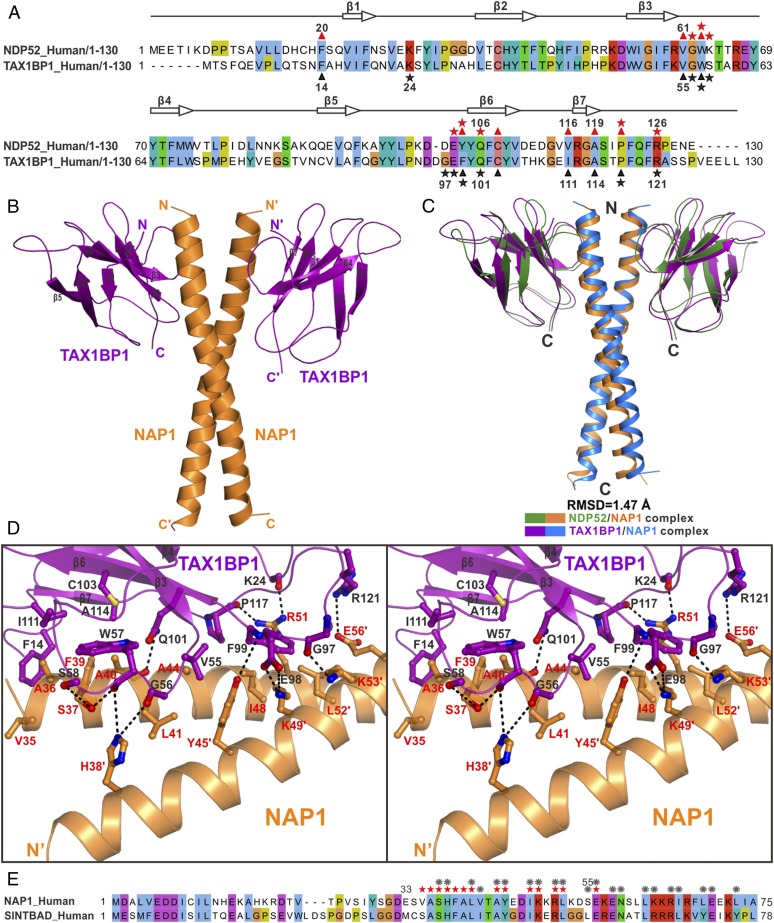
Fig. 5.
SKICH domains of TAX1BP1 and NDP52 share a general binding mode to interact with NAP1 and SINTBAD. (A) Structure-based sequence alignment analysis of the SKICH regions of NDP52 and TAX1BP1. In this alignment, the conserved residues are highlighted in colors, using Jalview2.8.1 software (www.jalview.org/). The key residues of NDP52 that interact with NAP1 are highlighted with red stars (polar interaction) or red triangles (hydrophobic interaction), and those of TAX1BP1 are highlighted in black. (B) Ribbon diagram showing the overall structure of TAX1BP1(1–121) in complex with NAP1(33–75). (C) Structural comparison of the overall structures of the TAX1BP1(1–121)/NAP1(33–75) complex and the NDP52(10–126)/NAP1(33–75) complex. In this drawing, the TAX1BP1(1–121) and NAP1(33–75) molecules in the TAX1BP1(1–121)/NAP1(33–75) complex are shown in purple and blue, respectively, while the NDP52(10–126) and NAP1(33–75) in the NDP52(10–126)/NAP1(33–75) complex are drawn in green and orange, respectively. (D) Stereoview of the ribbon-stick representation showing the detailed binding interface of the TAX1BP1/NAP1 complex. The related hydrogen bonds and salt bridges involved in the TAX1BP1 and NAP1 interaction are shown as dotted lines. (E) Sequence alignment analysis of the NDP52-binding regions in NAP1 and SINTBAD showing that most key interface residues of NAP1 that are crucial for dimerization or interacting with NDP52 are also conserved in SINTBAD. The interface residues of NAP1, which are critical for the interactions with NDP52 and/or TAX1BP1, are highlighted with red stars, while the NAP1 residues that are involved in the dimerization of NAP1 are labeled with gray gears.