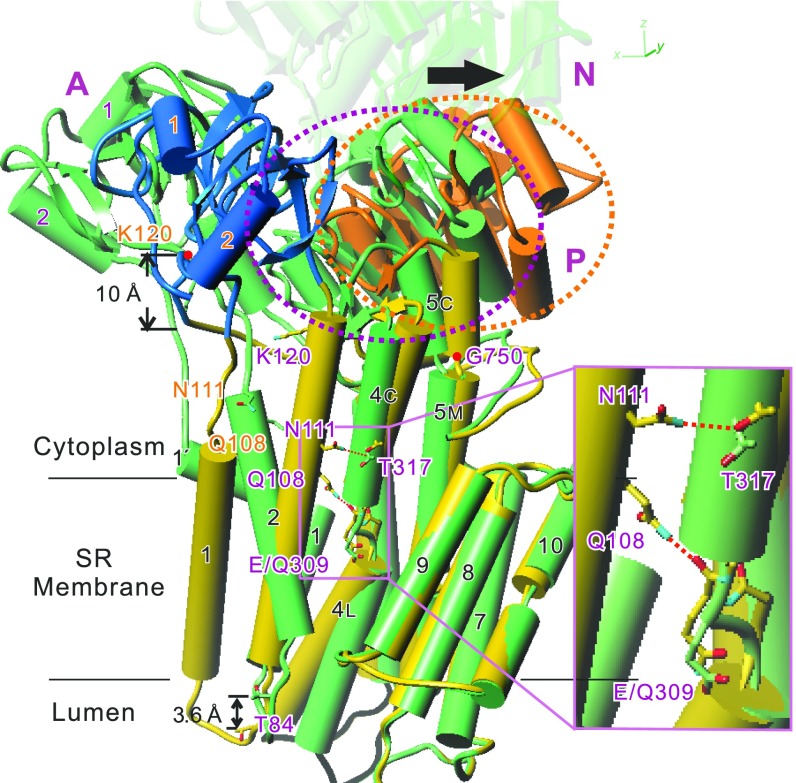
Fig. 2.
Superimposition of atomic models for the Glu309Gln mutant and native SERCA1a in E2(TG). Aligned with the M7-M10 helices. M6 is removed for clarity. The N-domain of Glu309Gln(TG) is removed and that of native ATPase is made transparent. The model for Glu309Gln(TG) appears in yellow with the A-domain in blue, the P-domain in orange, and native SERCA1a in green. Orange and purple dotted circles enclose the P-domain of Glu309Gln(TG) and native ATPase, respectively. M5 is a contiguous helix, but shown with three cylinders (M5C, M5M, and M5L). Note that hydrogen bonds are formed in Glu309Gln(TG) between the side chains of Asn111 and Thr317 and between the side chain of Gln108 and the main chain carbonyl of Gln309 (pink dotted lines). Also note that the downward shift of M2 is large (∼10 Å at Lys120) at the cytoplasmic end but reduced to ∼4 Å (at Thr84) at the other end due to winding of the M2 helix in the mutant. The arrow shows the movement of the P-domain (dotted circles) caused by the Glu309Gln substitution. Inset shows a magnified view of the boxed area.