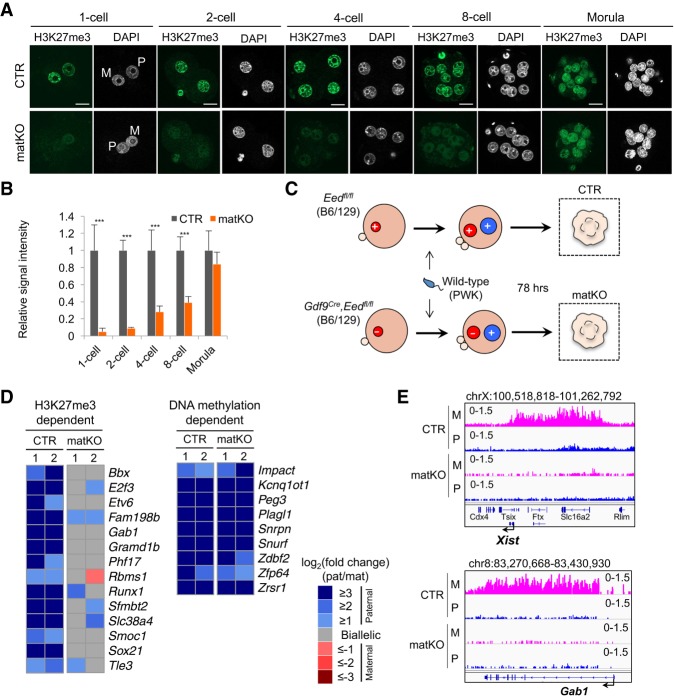
Figure 1.
Maternal Eed knockout induces loss of H3K27me3 imprinting. (A) Representative images of zygotes and preimplantation embryos immunostained with an anti-H3K27me3 antibody. (M) Maternal pronucleus; (P) paternal pronucleus. Bar, 20 µm. (B) Quantification of the H3K27me3 signal intensity. The average signal intensity of CTR embryos was set as 1.0. The total numbers of embryos examined were eight CTR and 12 matKO for one-cell embryos, six CTR and five matKO for two-cell embryos, seven CTR and six matKO for four-cell embryos, nine CTR and nine matKO for eight-cell embryos, and 10 CTR and eight matKO for morula embryos, respectively. Error bars indicate SD. (***) P < 0.001, two-tailed Student t-test. (C) Scheme for assessing allele-specific gene expression and H3K27me3 enrichment in morula embryos. (+) Eed wild-type allele; (–) Eed knockout allele. (D) Heat map showing the allelic expression bias of H3K27me3-dependent and known DNA methylation-dependent PEGs in morula embryos with biological duplicates. Genes with >20 single-nucleotide polymorphism (SNP) reads in both replicates are shown. (E) Genome browser views showing loss of maternal H3K27me3 domains at the representative H3K27me3-dependent imprinted loci in CTR and Eed matKO morula embryos. (P) Paternal allele; (M) maternal allele.