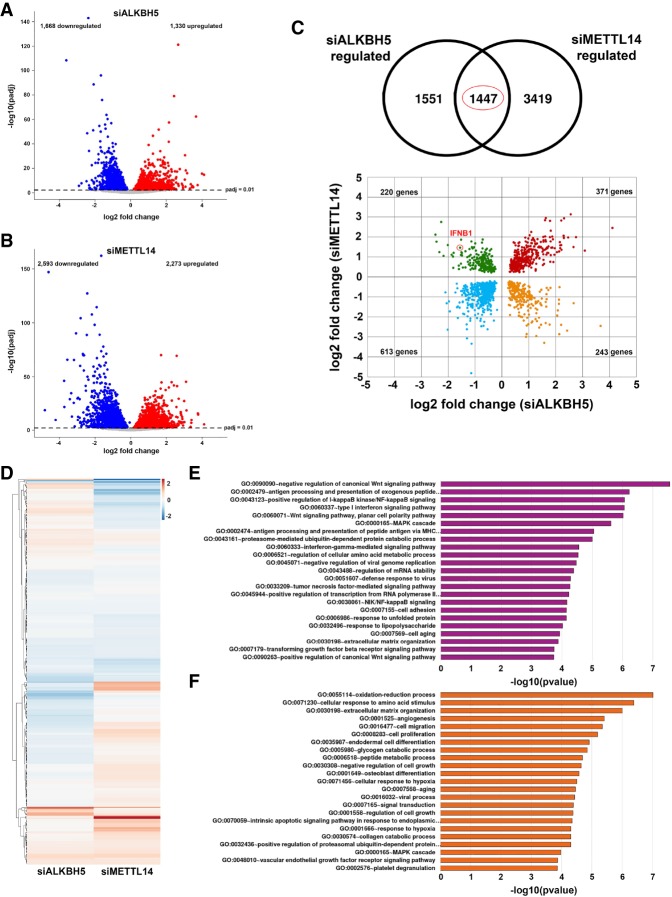
Figure 4.
Control of genome-wide responses to dsDNA by m6A demethylase ALKBH5 and m6A methylase subunit METTL14. Volcano plots show differentially expressed genes (adjusted P-value < 0.01) identified from RNA-seq of polyadenylated RNA collected from cells transfected with ALKBH5 siRNA (siALKBH5; n = 3 biological replicates) (A) or METTL14 siRNA (siMETTL14; n = 3 biological replicates) (B) and stimulated with dsDNA for 12 h. Genes up-regulated versus a nonsilencing siRNA control (stimulated with dsDNA for 12 h; n = 3 biological replicates) are shown in red, while down-regulated genes are shown in blue. Nonregulated genes are shown in gray. (C) One-thousand-four-hundred-forty-seven significantly regulated genes (P < 0.01) overlap between data sets generated using siALKBH5 and siMETTL14. A scatter plot of these shows genes that are commonly up-regulated or down-regulated following either METTL14 or ALKBH5 silencing (highlighted in red and blue, respectively). Genes regulated in a reciprocal manner are highlighted in green (up-regulated when METTL14 is silenced and down-regulated when ALKBH5 is silenced) and yellow (down-regulated when METTL14 is silenced and up-regulated when ALKBH5 is silenced). The red circle highlights the IFNB1 mRNA. (D) Heat map depicting 349 ISGs, colored according to log2 fold change in expression versus the nonsilencing siRNA control. (E,F) Pathway analyses (gene ontology direct terms) of significantly differentially expressed genes from A and B were conducted using DAVID and filtered according to a Benjamini-Hochberg procedure (<0.05).