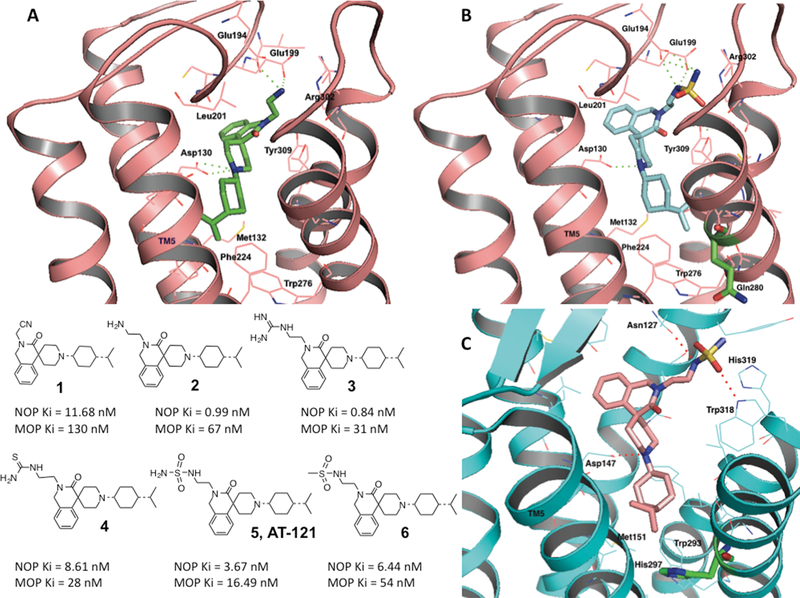
Figure 1. Structure-based design and optimization of bifunctional NOP-MOP ligands.
(A) top: Docking and binding interactions of compound 2 (shown as green stick representation) into the active-state NOP homology model (38), shown in cartoon representation (pink). Amino acids in a 4 angstrom radius around the ligand are shown as pink sticks and are labeled. Polar interactions of the 2-aminoethyl moiety of 2 with Glu194 and Glu199 in the extracellular loop (EL) 2 of the receptor are shown as green dotted lines. Bottom: structures of the isoquinolinone-based bifunctional ligands and their binding affinities at NOP and MOP. (B) Docking and binding interactions of compound 5 (AT-121) (shown in cyan) in the active-state NOP structure (pink). Note the interactions of the nitrogens of the sulfamide group with the Glu194 and Glu199 of the EL2 loop of NOP. Gln280, the non-conserved amino acid between NOP and the opioid receptors (His, shown in Fig. 1C), shown as green sticks, does not interact with the ligand. (C) Docking and binding interactions of bifunctional compound 5 in the MOP active-state crystal structure (73), shown in cyan cartoon representation. The interacting amino acids are shown as cyan stick representations. The lipophilic isopropylcyclohexyl group binds to a nonpolar pocket lined with Met131, His 297 (green sticks) and Trp 293 in the MOP binding pocket.