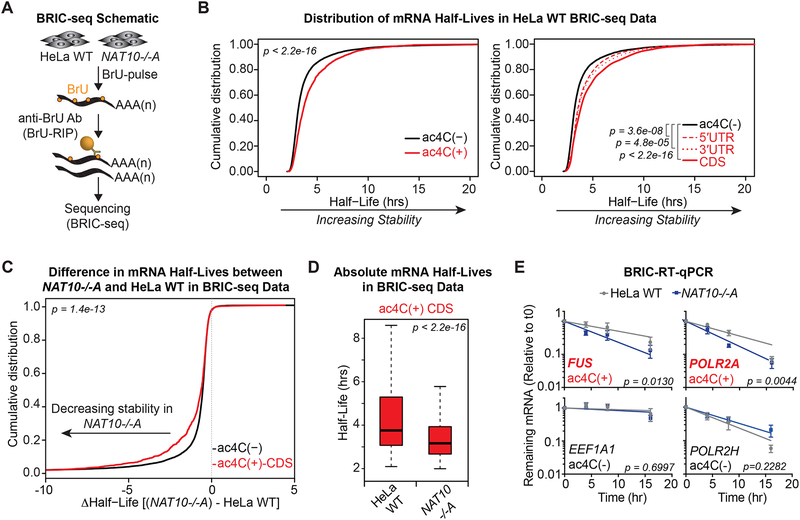
Figure 5. ac4C promotes mRNA stability.
(A) Schematic of 5’-bromo-uridine [BrU] immunoprecipitation chase-deep sequencing (BRIC-seq).
(B) Cumulative distribution plots of mRNA half-lives in parental HeLa cells for ac4C(−) (n=9,821) and all ac4C(+) (n=1,966) transcripts (left), or subdivided by ac4C summits occurring exclusively within 5’UTR (n= 248), 3’UTR (n=219), or CDS (n= 1,048) (right). p = KS test.
(C) CDF plot of differential mRNA half-lives in NAT10−/−A vs. parental HeLa cells for ac4C(−) and ac4C(+) transcripts with summit position within CDS. p = KS test.
(D) Boxplots of median half-lives of ac4C(+) transcripts with CDS summits in parental and NAT10−/−A HeLa cells. Boxes indicate median, 25th, and 75th percentiles, and whiskers extend to 1.5 times the interquartile range (excluding outliers). p = Wilcoxon rank-sum test.
(E) BrU-labeled RNA was immunoprecipitated as described in (A) followed by RT-qPCR. Decay graphs were generated by applying the One-Phase Decay model. Mean ± SEM, n=4, Sum-of-squares F test.