Figure 1. Targeting replication stress in glioblastoma stem-like cells (GSCs).
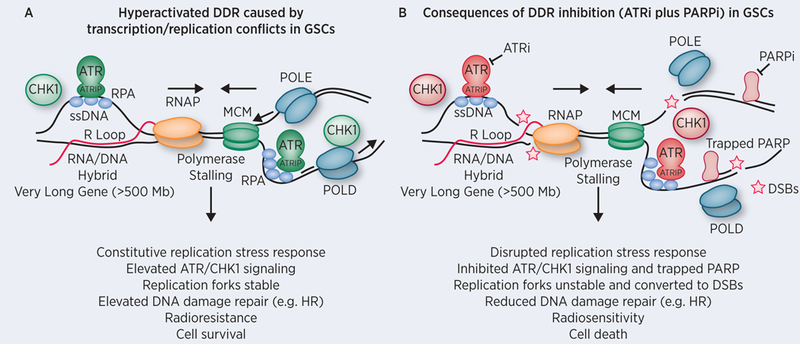
A) Carruthers et al. demonstrated that CD133+ GSCs exhibit constitutive replication stress (RS) as shown by elevated ATR and CHK1 kinase signaling (colored green). ATR is activated by binding to RPA coating extended regions of ssDNA through its partner ATR Interacting Protein (ATRIP). ATR phosphorylates and activates CHK1 thereby initiating a DNA damage response that promotes activation of the intra S and G2/M phase checkpoints, increases replication fork stability, and regulates DNA repair pathways such as homologous recombination (HR) (35). One potential source of RS in GSCs is the elevated transcription of ‘very long genes’ by RNA polymerase that may inadvertently collide with late replicating regions of the genome, activating the ATR replication stress response, which in turn promotes cell survival and radioresistance. B) Treatment of GSCs with an ATR inhibitor (colored red) is selectively toxic due to GSC dependence upon RS response signaling for survival. Inhibition of the RS response leads to increased R loop and replication fork instability that ultimately lead to DSBs following structure-specific endonuclease processing or DNA breakage. Inhibition of PARP results in base excision repair deficiency and may lead to trapping of the PARP enzyme on ssDNA breaks creating further dependence upon ATR signaling to promote stability and repair of stalled replication forks. Inhibition of ATR and PARP leads profound radiosenstization of GSCs. Abbreviations: ATR (Ataxia telangiectasia and Rad3-related), RPA (Replication Factor A), ssDNA (single-stranded DNA), RNAP (RNA polymerase II), MCM (minichromosome maintenance protein complex helicase), POLE and POLD (DNA polymerase epsilon and delta), PARP (Poly (ADP-ribose) polymerase), DSBs (Double-Stranded DNA breaks).
