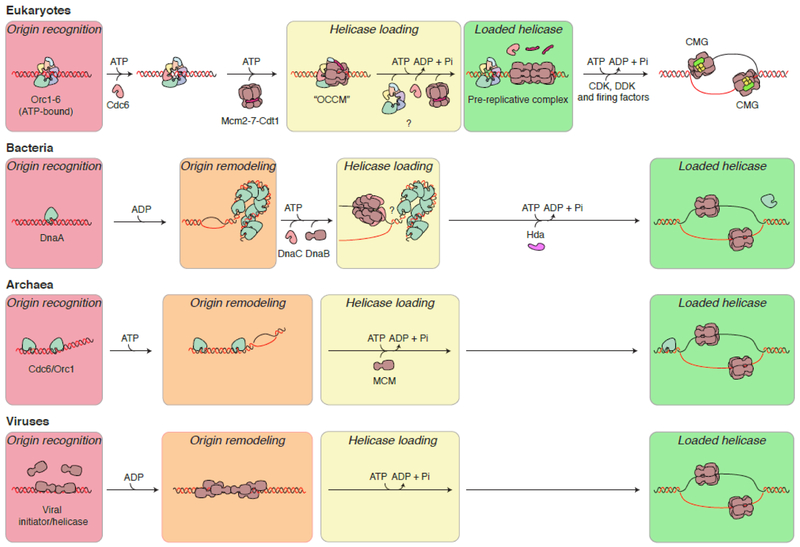
Fig. 1.
Models for origin recognition and helicase loading in the three domains of life and in viruses. In eukaryotes, bacteria and archaea, the origin is first recognized by the origin recognition machinery, with concomitant DNA remodeling occurring in bacteria and archaea. The replicative helicase is then recruited and directly interacts with the origin recognition complex. ATP binding but not hydrolysis is required for this interaction (see text). In eukaryotes, multiple origin recognition proteins may be required for loading a double hexameric form of the helicase. After helicase loading, ATP hydrolysis allows for reiterate assembly of helicases on other locations on the DNA. In viruses (human papillomavirus is shown as an example in this case), the viral helicase often serves the function of both origin recognition and self-assembly onto DNA into the mature helicase. In papillomavirus, a double-trimeric assembly of the helicase complex, linked to origin melting, is a prerequisite for the fully functional helicase complexes to be formed in subsequent steps in an ATP hydrolysis-dependent manner (for simplicity, other auxiliary factors are omitted in the diagram).