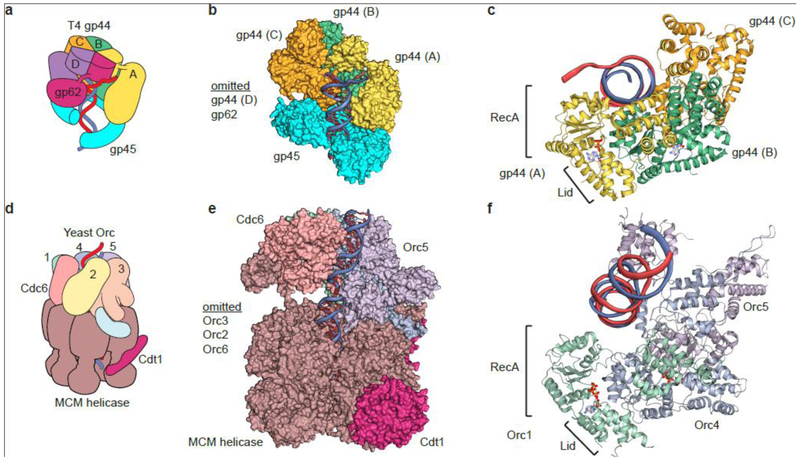
Fig. 4.
Comparison of loaders of ring-shaped proteins from bacteriophage T4 and S. cerevisiae.
(a) Schematic of the T4 DNA polymerase clamp loader (gp44 and gp62) bound to the gp45 clamp [an analogue of the eukaryotic PCNA]. (b) Surface representation of the structure of the T4 clamp loader bound to the clamp. (c) Cartoon representation of gp44 subunits A,B,C bound to primer-template DNA (red and blue strands) (d) Schematic of the S. cerevisiae helicase loader (ScORC+Cdc6) and pre-helicase complex (MCM+Cdt1). (e) Surface representation of the structure of ScOrc-Cdc6 bound to the MCM complex and Cdt1 on double strand origin DNA (f) Cartoon representation of ScORC subunits 1,4,5 bound to double-stranded DNA [red and blue strands]. (c) and (f) illustrate the corkscrew orientation of the subunits in the clamp loader and in ORC, both of which surround dsDNA.