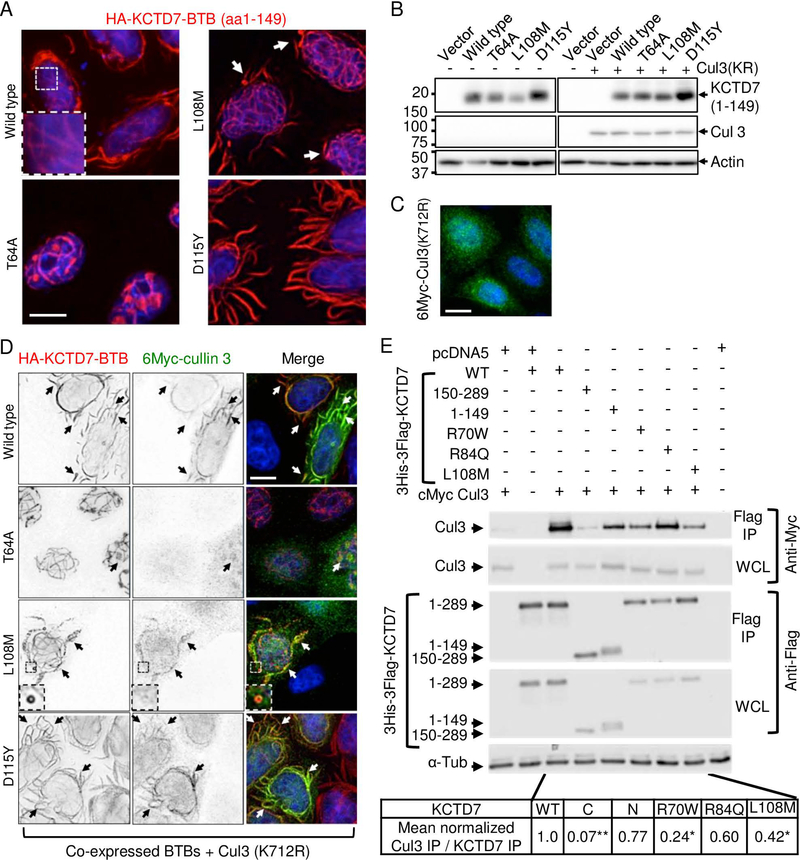
Fig 4. Patient mutations in the BTB domain affect cullin 3 (CUL3) interactions.
(A) Immunofluorescence microscopy of N-terminal HA-tagged KCTD7-BTB (amino acids 1–149) in Kyoto HeLa cells transfected 18 h and stained with 1:1000 anti-HA (Santa Cruz HA Y-11) and 1:2000 anti-Myc (Calbiochem Ab-1 OP10L) (similar results without tag and in other cell types tested). Representative of >3 independent experiments. (B) Immunoblots (12% SDS-PAGE, PVDF) of samples described for panels A, C and D probed for anti-HA (1:1000; Santa Cruz Y11), anti-Myc (1:2000; Calbiochem), and anti-actin as loading control (1:10,000; MP Biomedicals 691001), representative of two independent experiments each with both WT and inactive K712R mutant CUL3 yielding similar results. (C) Immunofluorescence microscopy of Kyoto HeLa cells transfected with N-terminal 6Myc-tagged CUL3 (K712R) alone and detected with anti-Myc (similar results for wild type 6Myc-CUL3). (D) Parallel samples to panels B and C co-transfected with HA-KCTD7(1–149) and N-terminal 6Myc-cullin3(K712R) and dual-stained with anti-Myc and anti-HA. Individual gray scale and color merged immunofluorescence microscopy images shown. Representative of >3 independent experiments per condition. Scale bar = 10 μm in all panels. (E) Co-immunoprecipitation of WT 6Myc-Cul3 from HEK293 whole cell lysates (WCL) after transient co-transfection with 3His-3Flag-KCTD7 using anti-Flag M2 affinity matrix for immunoprecipitation (IP). The strength of the interaction between CUL3 and KCTD7 variants was quantified as a ratio of the IP cMyc signal to the IP Flag signal and normalized to the WT KCTD7/CUL3 ratio in the total IP. Representative of 3 independent experiments is shown. *P<0.000001, *P<0.03