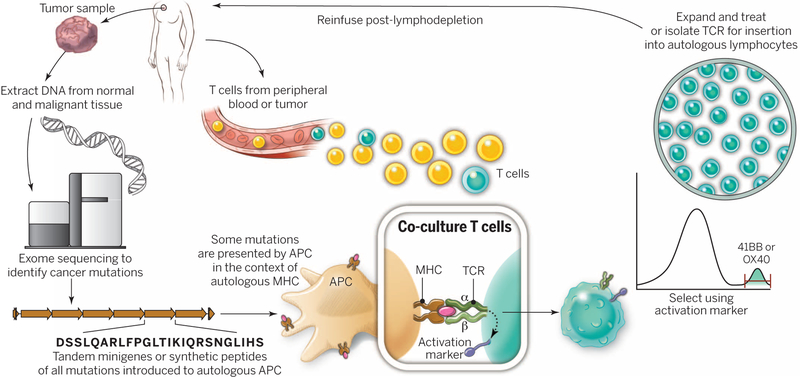
Fig. 3. A “blueprint” for the treatment of patients with T cells recognizing tumor-specific mutations.
The sequences of exomic DNA from tumor cells and normal cells from the same patient are compared to identify tumor-specific mutations. Knowledge of these mutations can then be used to synthesize either minigenes or polypeptides encoding each mutated amino acid flanked by 10 to 12 amino acids. These peptides or minigenes can be expressed by a patient’s autologous APCs, where they are processed and presented in the context of a patient’s MHC. Coculture of the patient’s T cells with these APCs can be used to identify all mutations processed and presented in the context of all of a patient’s MHC class I and class II molecules. The identification of individual mutations responsible for tumor recognition is possible because T cells express activation markers, such as 41BB (CD8+ T cells) and OX40 (CD4+ T cells), when they recognize their cognate target antigen. T cells expressing the activation marker can then by purified using flow cytometry before their expansion and reinfusion into the tumor-bearing patient.