Abstract
Adoptive transfer of receptor-engineered T cells has produced impressive results in treating patients with B cell leukemias and lymphomas. This success has captured public imagination and driven academic and industrial researchers to develop similar ‘off-the-shelf’ receptors targeting shared antigens on epithelial cancers, the leading cause of cancer-related deaths. However, the successful treatment of large numbers of people with solid cancers using this strategy is unlikely to be straightforward. Receptor-engineered T cells have the potential to cause lethal toxicity from on-target recognition of normal tissues, and there is a paucity of truly tumor-specific antigens shared across tumor types. Here we offer our perspective on how expanding the use of genetically redirected T cells to treat the majority of patients with solid cancers will require major technical, manufacturing and regulatory innovations centered around the development of autologous gene therapies targeting private somatic mutations.
Irrefutable evidence that an entirely immunologic approach can cause regression of a wide array of human cancers has come from the recent success of using monoclonal antibodies (mAbs) targeting checkpoints of immune activation, including cytotoxic T lymphocyte–associated protein 4 (CTLA-4) (ref. 1) and programmed cell death protein 1 (PD-1) (ref. 2). This includes patients affected with an ever-expanding list of malignancies, including melanoma1,2, renal cell carcinoma2,3, lung cancer2,4, bladder cancer5, ovarian cancer6, Hodgkin’s lymphoma7, and gastrointestinal (GI) and endometrial cancers associated with defects in DNA mismatch repair8. Despite different mechanisms of action, these immunotherapies culminate with the activation and expansion of tumor-reactive T cells9–12.
Because T cells are often are the final effectors of immune-mediated cancer regression, strategies that directly use tumor-reactive T cells as a therapy have been developed13. In this approach, termed adoptive cell transfer (ACT), T cells are expanded outside the potentially immunosuppressive environment of a tumor and re-infused in large numbers into the cancer patient (up to 1011 cells). Historically, procuring antitumor T cells for use in ACT has come from the surgical removal of a cancer metastasis in order to obtain tumor-infiltrating lymphocytes (TILs). TILs demonstrate tumor reactivity with variable frequency in a range of cancers, including melanoma14–17, GI18,19, lung20 and human papilloma virus–associated malignancies21. TIL infusion can induce durable complete responses (CRs)14,21, including in patients for whom other immunotherapies have failed14. Despite demonstrable efficacy, use of TIL outside the context of clinical trials performed at academic medical centers has proven challenging.
Progress in gene engineering technologies has simplified the generation of antitumor T cells, overcoming many of the practical barriers that have limited wide dissemination of ACT using TIL cells. Gene engineering obviates the requirement for surgery because T cells can be isolated from the blood and receptors conveying specificity for tumor-associated antigens can be introduced using viral and non-viral integration techniques22. Thus, antitumor T cells can potentially be made on a large scale using commercial production methods. Indeed, recent experience with sipuleucel-T, a gene-modified cell product for prostate cancer, demonstrated the feasibility of having a patient’s immune cells collected, sent to a central manufacturing facility, and returned back for re-infusion in a manner that gained US Food and Drug Administration (FDA) regulatory approval23. Finally, genetic modification of T cells has a track record of safety. Gammaretroviral and lentiviral vectors have been used most commonly in antigen receptor gene therapy trials. Despite concerns about the possibility of insertional mutagenesis24, introduction of antigen receptors into mature human T cells has been used to treat several hundred patients without evidence of clonal expansion or transformation25.
Collectively, a framework of manufacturing feasibility, regulatory precedent and vector safety is now in place and it is possible to envision treating large numbers of cancer patients using gene-engineered T cells. Recent success with gene-modified T cells targeting the B cell lineage differentiation antigen CD19 in a range of B cell malignancies has focused attention on using similar ‘off-the-shelf’ antigen receptors to treat patients with advanced solid cancers. In this Perspective, we offer our appraisal of how adoptive immunotherapy using receptor-engineered T cells can enter mainstream clinical oncology for patients with advanced epithelial cancers, the leading cause of cancer-related deaths26.
Antigen receptor–engineered T cells
T cell receptors.
Genetically redirecting a T cell’s specificity toward a patient’s cancer can be accomplished by the introduction of one of two types of antigen receptors. In one approach, a cloned T cell receptor (TCR) conferring tumor recognition is inserted into circulating lymphocytes. Similarly to the endogenous TCR expressed by all T cells, genetically introduced TCRs recognize a proteolytically processed peptide derived from either a cytosolic or membrane-associated protein presented within the groove of a specific major histocompatibility complex (MHC). Engineered TCRs trigger T cell activation through the signal transduction machinery used by the native TCR27. Thus, engineered TCRs are subject to the same counter-regulatory circuits that physiologically downregulate TCR signaling28,29.
Chimeric antigen receptors.
In a second approach, T cell specificity can be redirected by introduction of a synthetic recognition structure termed a chimeric antigen receptor (CAR). A CAR combines the antigen binding domain of a single-chain variable fragment (scFv) from a mAb that confers recognition of a tumor-associated antigen with intracellular signaling motifs capable of T cell activation30. In contrast to TCRs, CARs only recognize structures present on a cell’s surface, but this recognition occurs independently of a particular MHC molecule. MHC-independent antigen recognition enables CAR-modified T cells to treat any patient whose tumor expresses the target structure, and thus it potentially permits the treatment of tumors that have acquired defects in antigen processing and presentation31. Some CARs auto-signal, which leads to unrestrained cellular activation that results in apoptosis32, cytokine release independent of cognate antigens33, and immunologic exhaustion32,33. Expression of surface-inhibitory receptors such as PD-1 by CAR-modified T cells can attenuate their function, as it can with TCRs32,34.
Clinical trials.
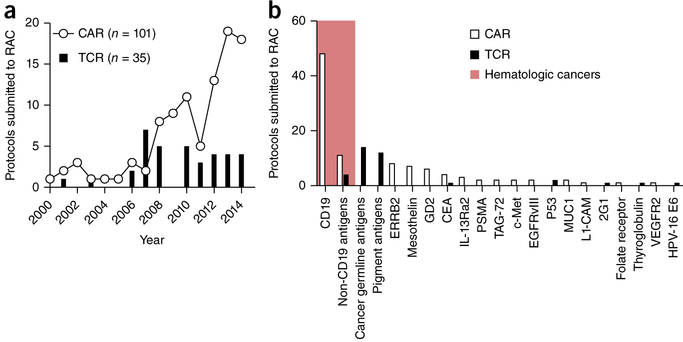
Since 1994, at least 148 human clinical trials have been initiated in the US for testing gene-engineered TCRs or CARs for the treatment of cancer (Fig. 1a) (ref. 35). Additional trials are also being conducted in Europe, Asia and Australia. Although the number of new TCR trials has remained relatively constant in recent years, the number of trials evaluating CAR-modified T cells has grown exponentially. In the majority of cases, the antigens targeted in these trials have been shared by tumors and normal tissues (Fig. 1b). Results from these trials have demonstrated that receptor-engineered T cells can, in some cases, mediate long-term remissions of selected solid and hematologic cancers.
Figure 1.
CAR and TCR clinical trials for oncology indications in the US between 1994 and 2014. (a) The number of new CAR (n = 101) and TCR (n = 35) clinical trials for hematologic and solid cancer indications submitted to the US Recombinant DNA Advisory Committee (RAC) as a function of time between 1994 and 2014. (b) Target antigens for CAR and TCR trials submitted to the RAC between 1994 and 2014. The shaded red area represents gene-engineered antigen receptor trials for hematologic cancers, the non-shaded area for solid cancers. Cancer-germline antigens include NY-ESO-1 and MAGE-A3, among others; pigment antigens include MART-1, gp100 and tyrosinase complex. ERRB2, HER-2/neu; CEA, carcinoembryonic antigen; IL-13Ra2, interleukin-13 receptor α2; PSMA, prostate-specific membrane antigen; TAG-72, tumor-associated glycoprotein 72; c-Met, MET proto-oncogene, receptor tyrosine kinase; EGFRvIII, epidermal growth factor receptor variant III; P53, tumor protein p53; MUC1, mucin 1, cell surface associated; L1-CAM, neural cell adhesion molecule L1; 2G1, a non-MHC-restricted TCR recognizing a TRAIL/DR4 complex; VEGFR2, vascular endothelial growth factor receptor 2; HPV-16 E6, human papillomavirus-16 E6 oncoprotein. Data is from the Genetic Modification Clinical Research Information System (http://www.gemcris.od.nih.gov).
For example, a recently completed TCR trial36 targeted a human leukocyte antigen (HLA)-A2–restricted epitope derived from NY-ESO-1, a cancer germline antigen (CGA) located on the X chromosome37. In this study, 11 of 20 patients (55%) with metastatic melanoma had objective evidence of cancer regression, including four CRs36. Importantly, three of these CRs were ongoing after >36 months. An additional 18 patients with synovial cell sarcoma, an aggressive connective tissue cancer associated with a characteristic t(X;18) chromosomal translocation37, were treated in this same study. Eleven of 18 (61%) treated patients had an objective response, including one CR. Although NY-ESO-1 is expressed by germ cells such as the testis, these tissues do not express MHC. Consequently, no on-target but off-tumor toxicities were observed.
A multitude of ACT studies have now demonstrated remarkable and frequently durable responses38 using CARs targeting CD19, a B cell–lineage antigen expressed on the surface of both normal B cells and many malignant B cells. After the first successful application of this approach in a follicular lymphoma patient39, CD19-specific CARs have been used effectively to treat patients with other chemotherapy-refractory B cell cancers including marginal zone lymphoma40, aggressive B cell lymphomas41, chronic lymphocytic leukemia38,40,42,43, and adult and pediatric acute lymphoblastic B cell leukemias (ALLs)44–48. Additionally, a recent case report suggested that a single patient with multiple myeloma had a sustained CR after an autologous stem cell transplant administered in conjunction with CD19-specific CAR cells49. This finding is currently being confirmed in a larger cohort of patients. Because CD19 is expressed by normal B cells, B cell aplasia and deficiencies in circulating immunoglobulins (Igs) are frequently observed in patients receiving CD19-specific CARs; these toxicities can be managed with Ig infusions. Many patients also exhibit cytokine release syndrome (CRS), a constellation of symptoms that occur after T cell infusion, and which is attributed to an exuberant release of cytokines, such as interferon (IFN)-γ and interleukin (IL)-6 (ref. 50). Symptoms associated with CRS include fevers, hypotension, hypoxemia, cardiac dysfunction, kidney failure and electrolyte abnormalities. Some patients also develop neurologic symptoms, including expressive aphasia, tremor and seizures, the cause of which remains unknown. In the majority of cases, these side effects can be managed with aggressive supportive care alone or in combination with immunosuppressants such as steroids or cytokine-specific antibodies. Nevertheless, treatment-related deaths have occurred at many institutions. Despite these complications, given the impressive clinical responses seen in patients with otherwise recalcitrant disease, we anticipate that CD19-specific CARs will enter mainstream care for many B cell malignancies in the next 1–2 years.
Limitations of current CAR and TCR approaches to treat common epithelial cancers
Two principles have emerged from successful TCR- and CAR- engineered ACT trials to date. First, potent antitumor effects in the absence of normal tissue damage can occur if the target is uniquely expressed by a patient’s tumor, as exemplified by the NY-ESO-1 TCR trials. Second, if a patient’s T cells are modified with a receptor that recognizes an antigen expressed both on non-transformed tissues and cancer cells, such as CD19 in the CD19-specific CAR trials, these cells will attack and destroy both normal and malignant tissue equally vigorously. Based on the success of the NY-ESO-1 TCR and the CD19-specific CARs, there is tremendous excitement in the immunotherapy field that similar ‘off-the-shelf’ TCRs and CARs targeting antigens shared across tumors, such as CGAs or tissue-differentiation antigens, will be highly effective against the majority of solid cancers.
Prerequisite to the generation of an antitumor CAR is knowledge of the genetic sequence from the scFv region of a mAb capable of recognizing antigens on a cancer cell’s surface. Although the testing of CARs in oncology clinical trials is comparatively new51,52, the search for tumor-specific antibodies is not53. Since the initial description in 1975 of a high-efficiency method for producing mAbs54, there has been a massive investment by academic and industry laboratories to develop therapeutic antibodies targeting cancer cells. Compared with the size of this investment, the search has yielded relatively few results.
To date, 20 mAb or mAb drug–radioisotope conjugates have been approved by the FDA for the treatment of cancer55. Among these, five target tumors indirectly by various mechanisms, including disruption of angiogenesis (bevacizumab, ramucirumab), interference with tumor-related bone remodeling (denosumab), or nonspecific immune activation through blockade of negative regulatory pathways such as CTLA-4 (ipilumimab) and PD-1 (nivolumab, pembrolizumab). For the remaining antibodies that directly target cancer cells, none are tumor-specific, but rather they recognize differentiation antigens also expressed by normal cells. Eight of these target lineage-specific antigens of the hematopoietic system, including CD20 (rituximab, ofatumumab, obinutuzumab, ibritumomab), CD30 (brentuximab vedotin), CD33 (gemtuzumab), CD38 (daratumumab) and CD52 (alemtuzumab) which are expressed on B cells, activated T and B cells, myeloid cells, lymphoid and myeloid cells, and lymphocytes, respectively56–60. Not unexpectedly, each of these antibodies induce cytopenias of benign cells that co-express these target antigens.
Of the remaining six mAbs that directly bind structures on the surface of solid cancers, five recognize growth factor receptors that are overexpressed by tumor cells, including epidermal growth factor receptor (EGFR, also known as ERB1; cetuximab, panitumumab) and ERBB2 (also known as HER-2/neu; trastuzumab, pertuzumab, ado-trastuzumab emtansine). For these mAbs, disruption of signal transduction pathways crucial to maintaining the malignant phenotype are the major, if not the only, mechanism of action55. Because EGFR and ERBB2 signaling is essential in the function of keratinocytes61 and cardiac myocytes62, mAbs targeting these receptors are associated with skin and cardiac toxicities. At present, the only example of an approved mAb that directly targets a solid-tumor antigen whose sole mechanism of action is immune-mediated killing is dinutuximab, which binds the glycolipid GD2 (ref. 63). Because GD2 is expressed not only by cancers, such as neuroblastoma and sarcoma, but also by peripheral sensory nerve fibers and neurons, neuropathic pain is a dose-limiting toxicity of this antibody63. Thus, after more than 35 years of clinical development, none of the approved mAbs that directly bind tumor cells are tumor specific, and all can mediate on-target and off-tumor toxicities.
CARs for solid cancers.
A similar situation appears to be the case for other tumor-targeted mAbs that are in clinical development and whose scFv regions are being incorporated into CAR designs. For example, the scFv of various antibodies targeting the shared tissue and tumor differentiation antigen mesothelin have been used to generate mesothelin-specific CARs for human clinical trials64,65. Mesothelin, a glycosylphosphatidyl inositol (GPI)-anchored membrane glycoprotein is highly expressed in a number of malignancies, including pleural and peritoneal mesothelioma, as well as pancreas, lung, breast, esophageal and ovarian cancer66. However, mesothelin is also expressed throughout the body in sensitive tissues that possess mesothelial cells. This includes the cornea, pleura, pericardium, peritoneum, tonsils, fallopian tubes and cervix. Consistent with this expression pattern, pleuritic-type chest pain was a dose-limiting toxicity in studies using mesothelin-specific antibodies conjugated to various immunotoxins66. Given the potential for substantial on-target toxicities, the FDA has required a protracted dose escalation in currently accruing phase 1 clinical trials testing the safety and efficacy of mesothelin-specific CARs. Recently, interim results from one mesothelin-specific CAR phase 1 study were presented in which as many as 3 × 107 cells were infused. Although no on-target toxicities were observed, neither was radiographic evidence of anti-tumor activity67. It is presently too early in the clinical development of mesothelin-specific CAR T cells to know whether mesothelin represents a viable target for gene-engineered T cells. Likewise, a recent CAR trial was initiated targeting MUC16 (ref. 68), a glycosylated mucin expressed on the surface of the majority of ovarian cancers, as well as in normal tissues that harbor mesothelial cells such as the eye69. As with the mesothelin-specific CAR trials, this study is also in the midst of a slow dose titration owing to safety concerns related to normal tissue targeting.
When sufficient data exists to assess the safety and efficacy of CAR-engineered T cells targeting other antigens shared by tumors and self tissues, substantial on-target toxicities have occurred. For example, CAR-modified T cells with an scFv specific for carbonic anhydrase IX (CAIX), an enzyme expressed by some kidney cancers and normal bile duct epithelial cells, triggered liver function abnormalities and cholangitis without causing cancer regression52. Similarly, infusion of ERBB2-specific CAR T cells constructed using the scFv from the humanized mAb trastuzumab resulted in lethal inflammatory cytokine release in the lung70. This toxicity was attributed to on-target but off-tumor recognition of low levels of ERBB2 expression on lung epithelial cells71. A more recent trial reported the administration of a separate ERBB2-specific CAR T cell in which the scFv was derived from FRP5, a mouse anti-human ERBB2-specific mAb72. Although no significant toxicities were reported, neither was evidence of in vivo CAR cell expansion or IFN-γ release in the blood after cell infusion. This might indicate that the engineered cells did not productively engage the target antigen. Although 1 of 19 patients treated with the FRP5-derived ERRB2-specific CAR had an objective anti-tumor response, this patient received salvage chemotherapy in addition to CAR T cell infusion. Thus, it is impossible to determine the relative contributions of the infused cells versus chemotherapy. These findings contrast with experience using CD19-specific CAR T cells in which cell expansion, cytokine release, and profound antitumor and on-target immunity against normal B cells is universally observed across trials40,41,43–48.
A possible exception to the paradigm of CAR targeting of a shared tumor and tissue differentiation antigen resulting in untenable on-target but off-tumor toxicities is GD2. In a series of pediatric patients with GD2-expressing neuroblastomas, infusion of a GD2-specific CAR resulted in tumor regression in 3 of 11 patients with active disease, including two sustained CRs73. No patients developed neuropathic pain, although several experienced somatic pain at the tumor site. These results are currently being confirmed in two clinical trials (NCT02107963 and NCT01822652) using a new GD2-specific CAR design.
Targeting of shared antigens.
As previously outlined in detail74, a similar pattern of on-target but off-tumor toxicities have been observed with gene-engineered TCRs targeting shared tumor and tissue differentiation antigens. This includes TCRs reactive against MART75, gp100 (ref. 75) and carcinoembryonic antigen (CEA)76. Collectively, these data suggest that targeting shared tissue differentiation antigens is likely to come with the price of toxicity to critical normal organs. Whereas toxicities related to CAR targeting of hematologic antigens such as CD19 are manageable with repletion therapies, the majority of solid tumors are not derived from ‘expendable’ or replaceable tissues.
One potential solution to minimizing undesired on-target but off-tumor toxicities is the engineering of safety and tissue-selectivity mechanisms into the transferred T cells (Box 1 and Fig. 2). A second solution to overcoming the limitation of on-target but off-tumor toxicity is the targeting of antigens uniquely expressed by tumor cells. The ideal antigen would be expressed in common across multiple tumor types and result from driver mutations that appear early in oncogenesis, directly contribute to the malignant phenotype, and are essential for cancer cell survival77. Examples could include epitopes encompassing hot-spot mutations in genes such as KRAS, NRAS, ALK, PI3K and BRAF, among many others. Similarly, epitopes derived from virally encoded genes, such as the human papillomavirus (HPV) E6 and E7 oncoproteins which cause cervical, anal, head and neck cancers, might also represent excellent targets because expression of these antigens is exclusive to cancer cells and not normal tissues74.
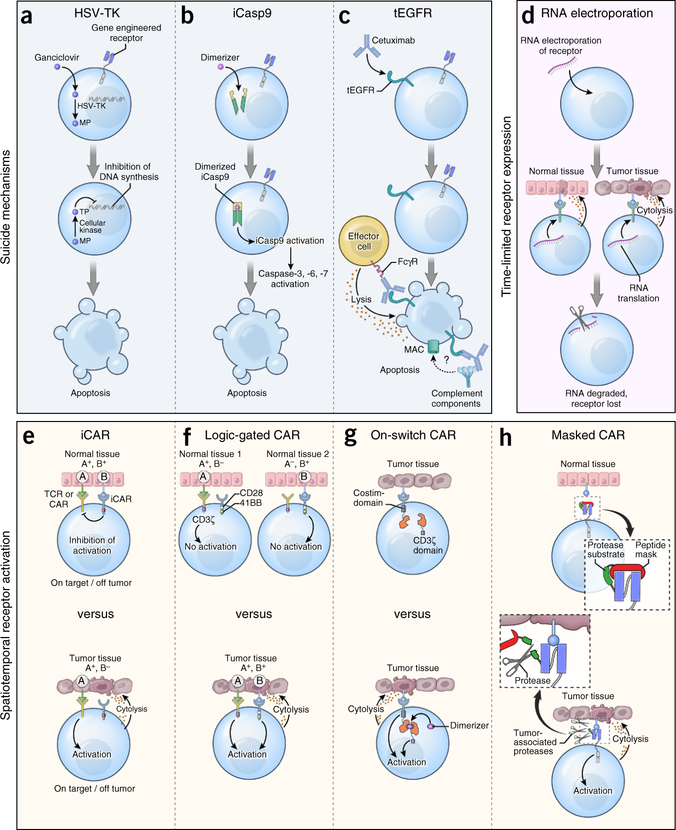
Box 1. Engineering safety and tissue-selectivity mechanisms into transferred T cells.
Complementing the identification of tumor-specific antigen receptors, strategies are being developed that enhance either the safety or tissue-selectivity of engineered T cells (Fig. 2).
Suicide genes.
T cells can be modified with a ‘suicide’ gene that confers sensitivity to a prodrug or antibody administered in the case of an adverse event. For example, insertion of herpes simplex virus–thymidine kinase (HSV-TK) renders T cells susceptible to the antiviral medication ganciclovir135. HSV-TK is the most extensively tested suicide gene in humans, and it was originally developed to modulate graft-versus-host disease (GvHD) after allogeneic transplantation135 or to attenuate immunopathology after ACT into immune-deficient hosts136. Because HSV-TK is a highly immunogenic virus-derived protein, cells expressing it can be immunologically rejected thereby compromising cellular persistence136. Alternative cell-suicide strategies have been developed using constructs derived from human proteins to reduce the risk of immune depletion. In the inducible caspase-9 (iCasp9) system, the sequence of human caspase-9 deleted for its endogenous activation domain is grafted onto a modified variant of human FK506-binding protein137. This allows for dimerization and activation of apoptosis upon ligation with a dimerizer drug. The in vivo activity of iCasp9 has been confirmed in patients with acute GvHD after allogeneic donor lymphocyte infusion138. Cells engineered with iCasp9 have been detected in patients for over 2 years after transfer139, indicating that the construct is not overtly immunogenic. Truncated EGFR (tEGFR) is another suicide gene derived from human sequences that uses the clinically approved EGFR-specific mAb cetuximab to deplete transduced cells140. The tEGFR molecule consists of an extracellular portion of human EGFR containing the epitope recognized by cetuximab. In pre-clinical studies, cetuximab caused antibody-dependent cellular cytotoxicity of tEGFR+ T cells. Clinical trials incorporating tEGFR are currently enrolling patients.
Spatiotemporal control of receptor activation.
Strategies that control the duration, location and timing of engineered receptor activity might also enhance safety. RNA electroporation introduces receptors into T cells with self-limited expression, because RNA does not integrate into a cell’s genome and is an inherently short-lived molecule in vivo141. In the event of toxicity, receptor expression on transferred cells will extinguish spontaneously within several days. Attempts to control the activation of receptor-engineered T cells have also been made. In one design termed logic-gated CARs, T cells are co-transduced with two separate CARs: one that provides suboptimal activation when stimulated alone, and a second that recognizes a separate tumor-associated antigen, which provides a co-stimulatory signal142. Thus, only simultaneous ligation of both receptors will allow T cell activation, providing an additional degree of specificity. Alternatively, CARs delivering a dominant antigen-specific inhibitory signal, termed inhibitory CARs (iCARs), have also been explored143. The generation of an iCAR is accomplished by attaching the signaling domains of co-inhibitory receptors such as CTLA-4 or PD-1 to a scFv that recognizes structures on normal tissues. In vitro, iCARs suppress cytokine release, cytotoxicity and cellular proliferation after exposure to targets that co-express antigens recognized by both the stimulatory receptor and the iCAR. Recently, the ability to selectively enhance antibody binding within the tumor microenvironment has been demonstrated using pro-antibodies144. In this design, the antigen-binding domain of an antibody is sterically blocked by a substrate peptide cleaved in the presence of matrix metalloproteinases enriched within the tumor microenvironment. Provided that the rate of CAR T cell recirculation is minimal once a receptor is unblocked, the use of these ‘masked’ CARs provides another means of focusing the activity of gene-engineered cells toward targets shared with healthy tissues. Finally, ‘on-switch’ CARs have been developed145. Here, the antigen-binding and intracellular-signaling domains of a receptor are separated into two components that assemble only in the presence of a small-molecule dimerizer. Thus, this system allows for pharmacologic control over CAR T cell activity. It is important to note that none of the spatiotemporal control mechanisms listed above have been tested in humans to date.
Figure 2.
Safety and tissue-selectivity mechanisms that may be inserted into gene-engineered T cells. (a) Co-expression of herpes simplex virus-thymidine kinase (HSV-TK) in antigen receptor-modified T cells. Following administration of the anti-viral medication ganciclovir (GCV), HSV-TK catalyzes the generation of GCV-monophosphate (MP) which is subsequently converted to GCV-trisphosphate (TP) by enzymes present in all mammalian cells. GCV-triphosphate inhibits DNA chain elongation in proliferating cells, resulting in lethal toxicity. (b) Co-expression of an inducible caspase-9 (iCasp9) construct in antigen receptor–modified T cells. Administration of a small molecule dimerizer induces dimerization and activation of iCasp9 that subsequently triggers executioner caspases-3, −6 and −7, resulting in apoptosis. (c) Co-expression of a truncated variant of human EGFR (tEGFR) in receptor-modified T cells. Infusion of the EGFR-specific mAb cetuximab results in antibody-dependent cellular cytotoxicity of tEGFR+ T cells once the antibody’s Fc domain is engaged by Fc gamma receptors (FcγR) on the surface of immune effector cells. Some reports also suggest that cetuximab might deplete tEGFR+ cells though complement fixation, but this mechanism of action remains controversial. MAC, membrane attack complex. (d) RNA electroporation of antigen receptors into T cells. Because of the short half-life of RNA species, receptor expression is self-limited after cell transfer, thereby restricting the potential for uncontrolled on-target but off-tumor toxicities. (e) CARs engineered to deliver a dominant antigen-specific inhibitory signal, termed inhibitory CARs (iCARs), work by attaching the signaling domains of co-inhibitory receptors to an antibody-binding region that recognizes structures on the surface of normal tissues. (f) CARs can be ‘logic-gated’ by co-transducing with two separate CARs: one that provides suboptimal activation when stimulated alone and a second that recognizes a separate antigen, which provides a co-stimulatory signal. Ligation of either receptor alone is insufficient to trigger T-cell activation whereas co-engagement allows T cells to proliferate, acquire effector functions, and exhibit on-target immunity only against tissues expressing both antigens. (g) CARs can be engineered with an ‘on-switch’, whereby the antigen-binding and cytosolic-signaling domains of the receptor are divided into distinct modules. Administration of a small-molecule dimerizer induces heterodimerization of these modules, initiating cellular activation only when both cognate antigen and the small molecule are present. Thus, the duration and intensity of receptor-engineered T cells can be controlled. (h) CARs can be engineered with a ‘masked’ receptor in which the antigen-binding domain is sterically blocked by a peptide mask attached to the receptor by a protease-cleavable linker. Entry of modified cells into tissues enriched in proteases, such as the tumor microenvironment, can cleave the blocking peptide and unmask the binding capacity of the CAR.
An example of a shared mutation in a solid cancer currently being targeted in immunotherapy clinical trials is EGFR variant III (EGFRvIII), a mutated version of EGFR resulting from an in-frame deletion of exons 2 to 7 of the gene78. This rearrangement leads to constitutive activation of the cell surface receptor and formation of an immunogenic epitope. Because EGFRvIII is expressed in approximately 30% of patients with glioblastoma multiforme, the most common and deadly primary adult brain tumor, it could represent an ideal immunotherapy target. Ongoing trials are testing the antitumor activity and safety of 2 different anti-EGFRvIII CAR designs79,80; however, it is currently too early to know whether these receptors will have clinical activity. Similarly, a TCR trial targeting an A2-restricted epitope derived from the E6 oncoprotein of the high risk HPV-16 serotype has also been initiated in patients with HPV-16+ malignancies81.
Another example of a group of antigens potentially shared across multiple tumors and not present on the surface of normal tissues are the CGAs. As noted above, the CGAs are a group of more than 100 potentially immunogenic proteins encoded by non-mutated genes whose expression in adult tissues is typically restricted to non-MHC-bearing germ cells37. During the genetic and epigenetic dysregulation leading to oncogenesis, CGA expression may be reactivated causing expression in a variety of cancer types. Most studies evaluating the frequency of CGA expression in cancer have used mRNA-based assays, such as quantitative PCR (qPCR). By using this technique, expression estimates approaching 50% or higher for patients with a given tumor type have been reported for many common epithelial malignancies, including bladder, esophageal, hepatocellular, non-small-cell lung and gastric cancers37,82. However, qPCR does not assess the uniformity of CGA expression among individual tumor cells within a patient. When protein expression techniques such as immunohistochemical (IHC) staining are used, strong expression of CGAs such as MAGE and NY-ESO-1 is often limited to only small numbers of cancer cells37,83. For example, in the original description of NY-ESO-1 expression by IHC83, only 2 of 13 tumors that showed expression as measured by qPCR also demonstrated protein expression in >50% of cancer cells, a finding confirmed by others37. This stands in sharp contrast to what is typically observed with tissue differentiation antigens, in which antigen expression is both more common and more pervasive84. From an immunotherapy perspective, treating patients with heterogeneous expression of a target antigen raises concerns about applying selection pressure for antigen-negative tumor cells. Indeed, in all published clinical trials in which objective clinical responses were observed after ACT of cells targeting CGAs, the patients had cancers with a high intensity of antigen expression in the majority (>50%) of cancer cells36,85,86. Whether ACT targeting a single antigen expressed on ≤50% of tumor cells can cause cancer regression is the subject of current pre-clinical and clinical investigations.
It is important to note that despite their name and the absence of off-target toxicities in trials testing an A2-restricted NY-ESO-1 TCR36,87, not all CGAs are exclusively cancer or germline specific. PRAME (preferentially expressed antigen in melanoma), for example, is a CGA with well-documented expression in healthy tissues such as the adrenal glands, placenta and endometrium88. Further, whereas the CGA MAGE-A3 appears not to be expressed in healthy tissues, MAGE-A12 and possibly other MAGE family members are expressed at low levels in selected areas of the brain86. This became tragically apparent when patients received ACT using T cells modified with an HLA-A2–restricted, affinity-enhanced MAGE-A3 TCR that cross-reacted with a non-identical A2-restricted MAGE-A12 epitope89. Among nine treated patients, four developed neurologic toxicities, including two neurologic-related deaths86. In a separate trial testing the efficacy of an HLA-A1–restricted, affinity-enhanced MAGE-A3-specific TCR, two cardiovascular-related deaths occurred90. Although MAGE-A3 expression was not detected in the heart, engineered T cells were found to cross-react with an unrelated peptide derived from the muscle-specific protein titin91.
Further limiting the number of potential patients who might benefit from TCR targeting of CGAs is the requirement that patients possess a HLA haplotype compatible with the TCRs available. Given that the most common MHC class I restriction elements used for TCR gene therapy studies, namely HLA-A1 and A2, are—at most—present in only about 15–50% of patients92, the number of patients who would be candidates for these TCR-engineered cells becomes incrementally smaller. Taken together, these data suggest that although TCR-engineered cells targeting certain CGAs can cause pronounced tumor regression without off-target toxicity in the subset of patients whose tumor cells uniformly express the target antigen and who possess a requisite HLA haplotype, the majority of patients with common solid cancers will not be candidates for such therapies.
Targeting private somatic mutations with autologous gene therapy Evidence supporting immunogenic neoantigens.
After a decades-long effort by our group and others targeting shared tissue differentiation antigens with cancer vaccines93 and gene-engineered T cells74, cancer immunologists are reassessing which antigens are responsible for immune-mediated cancer regression. Increasing clinical evidence supports the hypothesis that immunogenic products of somatic mutations unique to each patient’s cancer—so-called neoantigens—are the relevant targets for successful immunotherapies13,94. Neoantigens may represent ideal targets because somatic mutations are central to the formation of most cancers; in other words, mutations may be functionally important to drive tumor growth and/or invasion. Further, neoantigens are exclusive to tumor cells, minimizing the risk of on-target, off-tumor killing of healthy tissue. Finally, because the mutations from which neoantigens are derived are somatic, the repertoire of TCRs expressed on T cell progenitors do not encounter neoantigens during thymic development and therefore should not be deleted by negative selection. Consequently, TCRs with high affinity for neoantigens may be present in the circulation. Several lines of evidence support these suppositions.
First, melanoma TILs mediate curative responses with minimal autoimmune sequelae14. By contrast, transfer of gene-engineered T cells expressing high-affinity receptors for pigment antigens resulted in suboptimal antitumor responses and severe on-target but off-tumor toxicities75. Thus, reactivity to targets besides differentiation antigens appears responsible for a substantial portion of melanoma TIL anti-tumor efficacy. Consistent with this premise, high-throughput assays using large collections of MHC multimers loaded with shared antigens revealed a low frequency of TILs expressing TCRs that bind to pigment and cancer-germline antigens95. By contrast, TILs that release inflammatory cytokines in response to patient-specific neoantigens can be detected at much higher frequencies in the majority of evaluated patients15,16. Second, it seems that immune checkpoint inhibitors are particularly effective in cancers with high burdens of somatic mutations. This includes diseases such as melanoma, which carries a high mutation load as a result of UV exposure10,96, tobacco-associated lung cancer11, bladder cancer5 and cancers arising in patients with defects in DNA mismatch-repair enzymes8. Indeed, in people with melanoma and lung cancer, the mutational burden and neoantigen load is highly correlated with clinical benefit from treatment with blocking antibodies specific to CTLA-4 and PD-1 (refs. 10,11,96). Third, the frequency of circulating neoantigen-specific T cells increased in responding patients following treatment with antibodies specific to CTLA-4 (refs. 9,10) or PD-1 (ref. 11) Finally, we recently demonstrated that a single infusion of a near-clonal population of neoantigen-specific CD4+ T cells resulted in prolonged tumor regression in a patient with cholangiocarcinoma18. Taken together, these data suggest that the isolation and re-infusion of neoantigen-specific T cells might be required to mediate tumor regression without inducing undesired on-target but off-tumor toxicities. If the TCRs necessary to induce cancer regression target neoantigens resulting from private somatic mutations unique to each patient’s cancer, is individualized gene-engineered T cell therapy possible? We believe that with proper investment and sufficient technologic and regulatory innovation, the answer can be yes. Further, as outlined below it may be possible to assemble all the necessary elements required to generate such an autologous TCR gene therapy solely from the peripheral blood (Fig. 3).
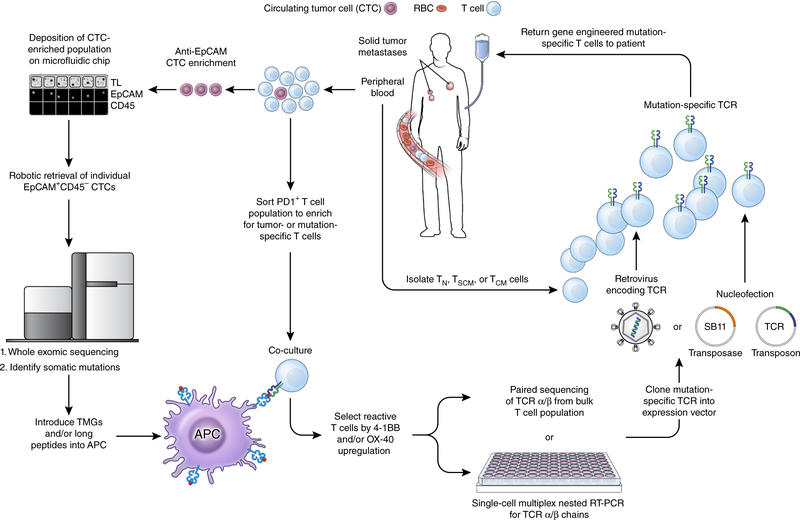
Figure 3.
A pathway for generating autologous TCR gene therapies targeting neoantigens for patients with advanced epithelial cancers. From a single blood draw, all of the requisite components required to produce this therapy can be procured. Circulating tumor cells (CTCs) can be enriched from the blood using a combination of antibody-mediated isolation based on epithelial marker expression (for example, EpCAM) followed by microfluidic isolation. Subsequent genomic extraction, amplification and whole exome sequencing can identify non-synonymous mutations present within the tumor. Circulating T cells that express PD-1 can be isolated and co-cultured with autologous professional antigen presenting cells (APCs) that have either been pulsed with synthetic long peptides harboring the amino acid change resulting from the mutation or RNA-electroporated with tandem minigenes (TMGs) encoding the amino acid change and flanked on either side by 12 amino acids from the endogenous protein. T cells that upregulate the activation markers 4–1BB and/or OX-40 can then be isolated and the α and β chains of the TCR associated with this cell can be sequenced. The α/β TCR that confers reactivity against a neoantigen can then be cloned into an expression vector, for example an integrating virus or the Sleeping Beauty (SB) transposon/transposase system. T cell subsets isolated from the peripheral blood of the patient can finally be modified with this expression vector, expanded in vitro to numbers sufficient for treatment, and re-infused back into the patient. TL, transmitted light; TN, T naive; TSCM, T stem cell memory; TCM, T central memory; SB11, SB transposase 11.
Identifying neoantigens and neoantigen-reactive T cells.
Currently, elucidation of the mutational landscape of a patient’s tumor as a first step in the detection of neoantigen-specific T cells is accomplished by performing whole-exome sequencing (WES) and/or RNA sequencing (RNA-seq) on tumor cells obtained through a tissue biopsy or surgical resection of a metastatic focus15,16,18,19. However, it is now possible to perform WES on circulating tumor cells (CTCs)97 or the cell-free products of tumor cells present in the circulation, such as tumor DNA98. These ‘liquid biopsies’ not only obviate the requirement for a patient to undergo an invasive procedure, they also can detect mutations held in common between the primary tumor and metastatic deposits97. Such founder or ‘trunk’ mutations are likely to be expressed in all tumor cells and therefore would make excellent immunotherapy targets. That said, not all detected mutations will be immunologically recognized as a cancer antigen. For a mutation to be immunogenic, it must be contained in an expressed gene, include a change in amino acid sequence resulting from either a nonsynonymous substitution or translocation, and be processed intracellularly into a 9- to 11–amino acid peptide capable of binding to one of the patient’s MHC molecules. Therefore, high-throughput methods for screening and isolating neoantigen-reactive T cells after a patient’s tumor has been sequenced are required. In one approach, synthetic long peptides that incorporate the substituted amino acid from a nonsynonymous mutation and that are predicted to be strong binders to one or more of the patient’s MHC molecules are pulsed onto autologous antigen-presenting cells (APCs)15,16. In a second approach, a minigene encoding the mutated amino acid and flanked on either side by 12 amino acids from the endogenous protein can be electroporated into autologous APCs17–19. With either technique, establishment of a patient-specific tumor cell line is not required.
One can use different sources of T cells to screen for neoantigen-reactive T cells. If a patient is able to undergo a metastatic resection, TILs can be isolated from this specimen, expanded and used as the screening population. Neoantigen-reactive TILs upregulate expression of tumor necrosis factor (TNF) family co-stimulatory markers such as 4–1BB and OX-40 (refs. 15,18,19,99,100), and/or the degranulation marker CD107a (ref. 100), when co-cultured with antigen-bearing APCs or autologous tumor cell lines. Such receptor upregulation can be used to identify neoantigen-specific T cells and to isolate these cells (either by fluorescence-activated cell sorting (FACS) or magnetic bead isolation) for further analysis100,101. Similarly, although PD-1 is a negative regulator of T cell functions, expression of this molecule also marks TILs that react with neoantigens99. Therefore, isolation of PD-1+ TILs can also be used to enrich for neoantigen-reactive T cells. Detection of neoantigen-specific TILs using these techniques can be successful not only in cancers with high mutational loads, such as melanoma, but also in cancers harboring relatively few mutations. For example, we recently reported that neoantigen-specific T cells could be identified from the TILs of nine out of ten sequentially screened patients with various gastrointestinal malignancies19. None of these patients had mutations in DNA mismatch-repair enzymes nor did they possess a microsatellite- unstable phenotype associated with a high mutational burden. Critically, a neoantigen-specific T cell population could be identified in a patient with pancreatic cancer whose tumor harbored only ten somatic mutations. These data demonstrate that it is possible to consistently identify neoantigen-specific TIL cells in common epithelial cancers that are not hyper-mutated and that seem to be unresponsive to other forms of immunotherapy, such as checkpoint inhibitors8.
In addition to TILs, neoantigen-specific T cells can also be identified directly from the peripheral blood of a cancer patient. This can be accomplished using a high-throughput methodology based on peptide-MHC complexes9,11,102. In this approach, proteasomal processing and HLA-binding algorithms are used to determine candidate epitopes containing nonsynonymous mutations predicted to bind with high affinity to one of the patient’s MHC alleles. Predicted peptides are subsequently synthesized and peptide-MHC complexes can be generated by UV-induced peptide exchange reaction103. By using this technique, it was shown in a recent report that it was possible to reproducibly identify, isolate and expand neoantigen-specific T cells from starting frequencies as low as 0.002% (ref. 102). Complementing this approach, it is probable that PD-1 may also identify tumor- reactive T cells in the circulation, just as it enriches for mutation-specific T cells in TILs99.
Engineering neoantigen-reactive T cells.
The mere isolation of mutation-specific and tumor-reactive T cells does not necessarily lead to an effective cancer treatment. This is because expansion of small numbers of isolated T cells to therapeutic numbers can result in a loss of replicative capacity and entry into a terminally differentiated state104. The development of effective T cell therapies from one or a limited number of cells might be possible with cellular reprogramming techniques105. The use of induced pluripotent stem cell (iPSC) technology with T cells has been experimentally accomplished106,107. Importantly, reprogrammed T cells retain a rearranged antigen-specific TCR. Although early reports suggested that iPSC-derived cells may be at increased risk for immunologic rejection after transfer into syngeneic hosts108, these findings have not been universally observed109,110. Other technical and practical hurdles remain in realizing the clinical utility of such T cell reprogramming techniques, however. For example, the efficiency of reprogramming remains low111, reprogrammed T cells can have an innate-like CD8αα or γδ T cell phenotype107,112, and differentiated cells derived from induced pluripotent cells might be at increased risk of undergoing malignant transformation113. Addressing each of these issues remain brisk areas of investigation.
An alternative approach using currently available technologies to generate minimally differentiated antitumor T cells could be cloning mutation-specific TCRs followed by inserting these TCRs into selected T cell subsets. This can be accomplished using high-throughput TCR gene-capture100 or multiplex nested single-cell real-time PCR (RT-PCR)114. More recently, it has been possible to perform α/β paired TCR sequencing (pairSEQ) from bulk populations of T cells using combinatorics115. After a mutation-specific α/β TCR has been identified, it subsequently can be cloned into a good manufacturing process (GMP)-quality expression system. In this case, substantial innovations in regulatory oversight are required.
It presently costs as much as $250,000 and a takes a minimum of 4–6 months to generate a GMP retroviral or lentiviral vector116. Both this expense and time scale are prohibitive for treating individual patients, although it might be feasible to simplify safety testing for human gene therapy trials117. A major contributor to the time and expense in generating a GMP viral vector is mandated biosafety testing. Whereas testing for sterility and the presence of mycoplasma is rapid and relatively inexpensive, testing for adventitious viruses, species-specific viruses, and replication-competent retrovirus (RCR) is not. Indeed, the necessity for routine RCR testing has been questioned by a large group of academic investigators involved in clinical gene therapy trials117. Given the potential for a favorable benefit/risk ratio in this patient demographic if autologous gene therapies are highly effective, it might be ethically permissible to relax some routine biosafety testing requirements. Ultimately, resolution of these issues will require close collaboration among all stakeholders, including regulatory agencies.
As an alternative to retroviral and lentiviral vectors, nonviral gene-transfer methods can potentially be used to genetically introduce antigen receptors. Because nonviral integration systems use oligonucleotides and recombinant proteins, they can be considerably cheaper to manufacture and easier to implement for single-use applications compared with viral vectors. By some estimates, production of nonviral reagents may cost one-tenth that of GMP-grade virus118. Presently, use of the Sleeping Beauty (SB) transposon/transposase system has advanced farthest in clinical development22. Genome editing strategies that introduce double-stranded DNA breaks that serve as sites for targeted gene insertion, including zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and the clustered regularly interspersed short palindromic repeat (CRISPR)-Cas9 system, might offer additional nonviral means of inserting antigen receptors in the near future119. Although the SB transposon/transposase system typically has a lower gene-transfer efficiency compared with viral integration techniques, selection for modified cells by drug exposure120, magnetic bead isolation121, or propagation using artificial antigen presenting cells122 can improve the frequency of receptor-engineered cells.
Finally, it will probably be advisable to enrich for ‘younger’ T cell subsets with high proliferative and engraftment potentials, such as naive, stem cell memory and central memory T cells, before introduction of the receptor104,123. In preclinical studies, these subsets are superior to more-differentiated effector memory and effector T cell subsets in mediating cancer regression124. Isolation of defined T-cell populations can be accomplished using antibody-microbead conjugates125 or streptamer126 isolation strategies. Each of these activities, including cell processing, genetic engineering and cell expansion, can be conducted at one of several centralized GMP cell manufacturing facilities within the US. These include facilities that were previously used to generate the sipuleucel-T product23, as well as newer facilities that are generating CD19-specific CARs.
Limitations targeting neoantigens.
Even with resolution of the technical and regulatory barriers listed above, critical questions regarding the targeting of neoantigens using patient-specific receptors remain. First, it is unclear how many receptors targeting distinct antigens might be required to reliably induce responses. Clinical experience using ACT of CD19-specific CAR- and NY-ESO-1–specific TCR-modified cells demonstrates that it is possible to induce durable CRs by targeting a single epitope36,38,41,47. However, this experience might not be transferable to the targeting of antigens generated by somatic mutations. Heterogeneity of the mutational landscape within a tumor mass and between metastases has been well documented in solid cancers127,128, raising the possibility that not all cancer cells within a single tumor or within a single patient will express the cognate target for a neoantigen-specific receptor. Under the selective pressure of an antigen-specific immune response, outgrowth of cancer cells lacking the target antigen might occur, a phenomenon recently documented in treatment failures with CD19-specific CAR cells129. Preclinical ACT studies in mice give conflicting conclusions as to whether direct cell killing of individual cancer cells expressing a target antigen is required to induce tumor regression, or whether bystander destruction is sufficient130–132. For example, epitope spreading has been proposed as a mechanism by which T cell killing of a limited population of tumor cells can lead to immunologic destruction of other tumor cells expressing unrelated antigens. There are several reported examples of patients receiving ACT in which transfer of a mono-specific T cell population resulted in increased immune reactivity against unrelated tumor or viral antigens85,133. Evidence for epitope spreading has also been documented in some patients responding to vaccine immunothera-pies134, however it is not known whether this phenomenon is causally associated with tumor regression. It would therefore seem wise to target multiple antigens using a panel of different receptors. Similarly, targeting neoantigens derived from the products of a driver or truncal mutation expressed in the primary tumor might reduce the risk of targeting a subpopulation of clonally divergent tumor cells.
Concluding remarks
Genetic redirection of patient T cells toward the B cell lineage antigen CD19 using CD19-specific CARs has met with unprecedented success in a heterogeneous group of chemotherapy-refractory hematologic diseases. So successful is this approach that it is very likely to become part of the standard of care for these diseases in the near future. Although this approach comes with the toxicity of normal B cell depletion, most patients can survive with Ig repletion. Nevertheless, the simple extrapolation of the experience using CARs in the treatment of lymphoid cancers to the treatment of most solid tumors is likely to come with the steep price of toxicity to essential normal tissues. Success for cell-based immunotherapies may come from the arduous task of targeting the unique set of mutations that cause each patient’s cancer.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research of the US National Institutes of Health (NIH) (ZIA BC011586 and ZIA BC010763) and the NIH Center for Regenerative Medicine. Additional support was provided by generous gifts from L. Jinyuan of the Tiens Charitable Foundation and the Milstein Family Foundation.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Hodi FS et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med 373, 1803–1813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer J et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powles T et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hamanishi J et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J. Clin. Oncol 33, 4015–4022 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Ansell SM et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med 372, 2509–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rooij N et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol 31, e439–e442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg SA & Restifo NP Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg SA et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res 17, 4550–4557 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linnemann C et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med 21, 81–85 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Robbins PF et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu YC et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin. Cancer Res 20, 3401–3410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran E et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science http://dx.doi.org/10.1126/science.aad1253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djenidi F et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J. Immunol 194, 3475–3486 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Stevanović S et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J. Clin. Oncol 33, 1543–1550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh H, Huls H, Kebriaei P & Cooper LJ A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol. Rev 257, 181–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantoff PW et al. IMPACT Study Investigators. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Hacein-Bey-Abina S et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest 118, 3132–3142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholler J et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med 4, 132ra53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel RL, Miller KD & Jemal A Cancer statistics, 2015. CA Cancer J. Clin 65, 5–29 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Ahmadi M et al. CD3 limits the efficacy of TCR gene therapy in vivo. Blood 118, 3528–3537 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Abate-Daga D et al. Expression profiling of TCR-engineered T cells demonstrates overexpression of multiple inhibitory receptors in persisting lymphocytes. Blood 122, 1399–1410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer DC et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J. Exp. Med 212, 2095–2113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eshhar Z, Waks T, Gross G & Schindler DG Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc. Natl. Acad. Sci. USA 90, 720–724 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khong HT & Restifo NP Natural selection of tumor variants in the generation of ‘tumor escape’ phenotypes. Nat. Immunol 3, 999–1005 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long AH et al. 4–1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frigault MJ et al. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol. Res 3, 356–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John LB et al. Anti-PD-1 antibody therapy potently enhances the eradication of established tumors by gene-modified T cells. Clin. Cancer Res 19, 5636–5646 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Recombinant DNA Advisory Committee Meeting Workshop on Cytokine Release Syndrome after T Cell Immunotherapy June 10th, 2015. https://auth.osp.od.nih.gov/sites/default/files/resources/RAC_Agenda_Day2%28CRS%29_UPDATED.pdf. 6-15-2015.
- 36.Robbins PF et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1–reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res 21, 1019–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caballero OL & Chen YT Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci 100, 2014–2021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter DL et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med 7, 303ra139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kochenderfer JN et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116, 4099–4102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochenderfer JN et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood 119, 2709–2720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kochenderfer JN et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol 33, 540–549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brentjens RJ et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118, 4817–4828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter DL, Levine BL, Kalos M, Bagg A & June CH Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brentjens RJ et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med 5, 177ra38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grupp SA et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med 368, 1509–1518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee DW et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maude SL et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davila ML et al. Efficacy and toxicity management of 19–28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med 6, 224ra25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garfall AL et al. Chimeric antigen receptor T cells against CD19 for multiple myeloma. N. Engl. J. Med 373, 1040–1047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee DW et al. Current concepts in the diagnosis and management of cytokine release syndrome. Blood 124, 188–195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kershaw MH et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin. Cancer Res 12, 6106–6115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamers CH et al. Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: first clinical experience. J. Clin. Oncol 24, e20–e22 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Rettig WJ & Old LJ Immunogenetics of human cell surface differentiation. Annu. Rev. Immunol 7, 481–511 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Köhler G & Milstein C Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 (1975). [DOI] [PubMed] [Google Scholar]
- 55.Redman JM, Hill EM, AlDeghaither D & Weiner LM Mechanisms of action of therapeutic antibodies for cancer. Mol. Immunol 67, 2 Pt A, 28–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stashenko P, Nadler LM, Hardy R & Schlossman SF Characterization of a human B lymphocyte-specific antigen. J. Immunol 125, 1678–1685 (1980). [PubMed] [Google Scholar]
- 57.Ellis TM, Simms PE, Slivnick DJ, Jäck HM & Fisher RI CD30 is a signal-transducing molecule that defines a subset of human activated CD45RO+ T cells. J. Immunol 151, 2380–2389 (1993). [PubMed] [Google Scholar]
- 58.Andrews RG, Torok-Storb B & Bernstein ID Myeloid-associated differentiation antigens on stem cells and their progeny identified by monoclonal antibodies. Blood 62, 124–132 (1983). [PubMed] [Google Scholar]
- 59.Malavasi F et al. Human CD38: a glycoprotein in search of a function. Immunol. Today 15, 95–97 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Rao SP et al. Human peripheral blood mononuclear cells exhibit heterogeneous CD52 expression levels and show differential sensitivity to alemtuzumab mediated cytolysis. PLoS One 7, e39416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miettinen PJ et al. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376, 337–341 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Crone SA et al. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat. Med 8, 459–465 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Yu AL et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med 363, 1324–1334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpenito C et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc. Natl. Acad. Sci. USA 106, 3360–3365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adusumilli PS et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci. Transl. Med 6, 261ra151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pastan I & Hassan R Discovery of mesothelin and exploiting it as a target for immunotherapy. Cancer Res 74, 2907–2912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanyi JL et al. Abstract CT105: Safety and feasibility of chimeric antigen receptor modified T cells directed against mesothelin (CART-meso) in patients with mesothelin expressing cancers. Proceedings of AACR Annual Meeting 2015. Cancer Res http://dx.doi.org/10.1158/1538-7445.AM2015-CT105 (2015). [Google Scholar]
- 68.Koneru M, O’Cearbhaill R, Pendharkar S, Spriggs DR & Brentjens RJ A phase I clinical trial of adoptive T cell therapy using IL-12 secreting MUC-16(ecto) directed chimeric antigen receptors for recurrent ovarian cancer. J. Transl. Med 13, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haridas D et al. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J 28, 4183–4199 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Morgan RA et al. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther 18, 843–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Press MF, Cordon-Cardo C & Slamon DJ Expression of the HER-2/neu proto-oncogene in normal human adult and fetal tissues. Oncogene 5, 953–962 (1990). [PubMed] [Google Scholar]
- 72.Ahmed N et al. Human epidermal growth factor receptor 2 (HER2) -specific chimeric antigen receptor-modified T cells for the immunotherapy of HER2-positive sarcoma. J. Clin. Oncol 33, 1688–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Louis CU et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood 118, 6050–6056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hinrichs CS & Restifo NP Reassessing target antigens for adoptive T-cell therapy. Nat. Biotechnol 31, 999–1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson LA et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parkhurst MR et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther 19, 620–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martincorena I & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Wong AJ et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Natl. Acad. Sci. USA 89, 2965–2969 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morgan RA et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Hum. Gene Ther 23, 1043–1053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson LA et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci. Transl. Med 7, 275ra22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Draper LM et al. Targeting of HPV-16+ epithelial cancer cells by TCR gene engineered T cells directed against E6. Clin. Cancer Res 21, 4431–4439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rosenberg SA Cell transfer immunotherapy for metastatic solid cancer--what clinicians need to know. Nat. Rev. Clin. Oncol 8, 577–585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jungbluth AA et al. Immunohistochemical analysis of NY-ESO-1 antigen expression in normal and malignant human tissues. Int. J. Cancer 92, 856–860 (2001). [DOI] [PubMed] [Google Scholar]
- 84.Chen YT et al. Serological analysis of Melan-A(MART-1), a melanocyte-specific protein homogeneously expressed in human melanomas. Proc. Natl. Acad. Sci. USA 93, 5915–5919 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hunder NN et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N. Engl. J. Med 358, 2698–2703 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morgan RA et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother 36, 133–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rapoport AP et al. NY-ESO-1–specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med 21, 914–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikeda H et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 6, 199–208 (1997). [DOI] [PubMed] [Google Scholar]
- 89.Chinnasamy N et al. A TCR targeting the HLA-A*0201-restricted epitope of MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen superfamily in several types of cancer. J. Immunol 186, 685–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Linette GP et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122, 863–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cameron BJ et al. Identification of a titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med 5, 197ra103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gonzalez-Galarza FF, Christmas S, Middleton D & Jones AR Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res 39, D913–D919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klebanoff CA, Acquavella N, Yu Z & Restifo NP Therapeutic cancer vaccines: are we there yet? Immunol. Rev 239, 27–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Andersen RS et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res 72, 1642–1650 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Van Allen EM et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lohr JG et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol 32, 479–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murtaza M et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Gros A et al. PD-1 identifies the patient-specific CD8 tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest 124, 2246–2259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Linnemann C et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat. Med 19, 1534–1541 (2013). [DOI] [PubMed] [Google Scholar]
- 101.Ye Q et al. CD137 accurately identifies and enriches for naturally occurring tumor-reactive T cells in tumor. Clin. Cancer Res 20, 44–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen CJ et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest 125, 3981–3991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodenko B et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc 1, 1120–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Gattinoni L, Klebanoff CA & Restifo NP Paths to stemness: building the ultimate antitumour T cell. Nat. Rev. Cancer 12, 671–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crompton JG, Clever D, Vizcardo R, Rao M & Restifo NP Reprogramming antitumor immunity. Trends Immunol 35, 178–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vizcardo R et al. Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8+ T cells. Cell Stem Cell 12, 31–36 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Nishimura T et al. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell 12, 114–126 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Zhao T, Zhang ZN, Rong Z & Xu Y Immunogenicity of induced pluripotent stem cells. Nature 474, 212–215 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Araki R et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 494, 100–104 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Guha P, Morgan JW, Mostoslavsky G, Rodrigues NP & Boyd AS Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 12, 407–412 (2013). [DOI] [PubMed] [Google Scholar]
- 111.Hanna JH, Saha K & Jaenisch R Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143, 508–525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Themeli M et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat. Biotechnol 31, 928–933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee AS, Tang C, Rao MS, Weissman IL & Wu JC Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med 19, 998–1004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang GC, Dash P, McCullers JA, Doherty PC & Thomas PG T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl. Med 4, 128ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Howie B et al. High-throughput pairing of T cell receptor α and β sequences. Sci. Transl. Med 7, 301ra131 (2015). [DOI] [PubMed] [Google Scholar]
- 116.Feldman SA et al. Rapid production of clinical-grade gammaretroviral vectors in expanded surface roller bottles using a ‘modified’ step-filtration process for clearance of packaging cells. Hum. Gene Ther 22, 107–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bear AS et al. Replication-competent retroviruses in gene-modified T cells used in clinical trials: is it time to revise the testing requirements? Mol. Ther 20, 246–249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Singh H et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 68, 2961–2971 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cox DB, Platt RJ & Zhang F Therapeutic genome editing: prospects and challenges. Nat. Med 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kacherovsky N, Liu GW, Jensen MC & Pun SH Multiplexed gene transfer to a human T-cell line by combining Sleeping Beauty transposon system with methotrexate selection. Biotechnol. Bioeng 112, 1429–1436 (2015). [DOI] [PubMed] [Google Scholar]
- 121.Field AC et al. Comparison of lentiviral and sleeping beauty mediated αβ T cell receptor gene transfer. PLoS One 8, e68201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rushworth D et al. Universal artificial antigen presenting cells to selectively propagate T cells expressing chimeric antigen receptor independent of specificity. J. Immunother 37, 204–213 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klebanoff CA, Gattinoni L & Restifo NP Sorting through subsets: which T-cell populations mediate highly effective adoptive immunotherapy? J. Immunother 35, 651–660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klebanoff CA et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin. Cancer Res 17, 5343–5352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Casati A et al. Clinical-scale selection and viral transduction of human naïve and central memory CD8+ T cells for adoptive cell therapy of cancer patients. Cancer Immunol. Immunother 62, 1563–1573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stemberger C et al. Novel serial positive enrichment technology enables clinical multiparameter cell sorting. PLoS One 7, e35798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yachida S et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sotillo E et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spiotto MT, Rowley DA & Schreiber H Bystander elimination of antigen loss variants in established tumors. Nat. Med 10, 294–298 (2004). [DOI] [PubMed] [Google Scholar]
- 131.Breart B, Lemaître F, Celli S & Bousso P Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J. Clin. Invest 118, 1390–1397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kerkar SP et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J. Clin. Invest 121, 4746–4757 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bollard CM et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J. Clin. Oncol 32, 798–808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Corbière V et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res 71, 1253–1262 (2011). [DOI] [PubMed] [Google Scholar]
- 135.Bonini C et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science 276, 1719–1724 (1997). [DOI] [PubMed] [Google Scholar]
- 136.Riddell SR et al. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat. Med 2, 216–223 (1996). [DOI] [PubMed] [Google Scholar]
- 137.Straathof KC et al. An inducible caspase-9 safety switch for T-cell therapy. Blood 105, 4247–4254 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Di Stasi A et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med 365, 1673–1683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou X et al. Long-term outcome after haploidentical stem cell transplant and infusion of T cells expressing the inducible caspase-9 safety transgene. Blood 123, 3895–3905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang X et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Y et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res 70, 9053–9061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kloss CC, Condomines M, Cartellieri M, Bachmann M & Sadelain M Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol 31, 71–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fedorov VD, Themeli M & Sadelain M PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med 5, 215ra172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Desnoyers LR et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med 5, 207ra144 (2013). [DOI] [PubMed] [Google Scholar]
- 145.Wu CY, Roybal KT, Puchner EM, Onuffer J & Lim WA Remote control of therapeutic T cells through a small-molecule-gated chimeric receptor. Science 350, aab4077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]