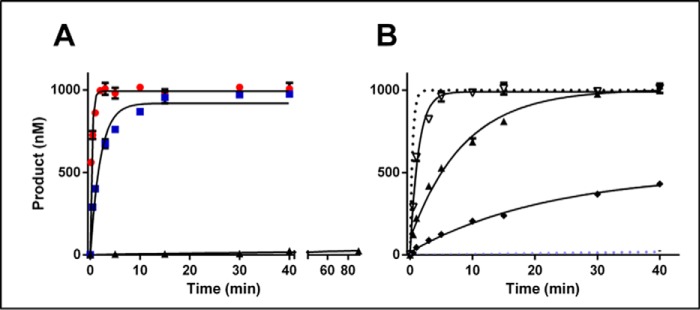
Figure 3.
The basis of RNA discrimination by CcrM C. crescentus. Single-turnover experiments with CcrM C. crescentus and selectively modified single-stranded 30-nucleotide substrates were performed. A, oligonucleotide made up exclusively of deoxyriboses displayed with red circles. The blue squares represent the reactivity of the substrate with three-flanking sugars on either side of the GANTC being ribose (5′-rArGrGGACTCrGrCrC-3′). The black triangles have riboses at all five internal positions (5′-AGGrGrArCrTrCGCC-3′). B, the DNA control is given for perspective as a dotted gray line (5′-AGGGACTCGCC-3′), and the all internal ribose is given as a dotted light blue line (5′-AGGrGrArCrTrCGCC-3′). The open upside-down triangles represent a single ribose substitution at the adenine (5′-AGGGrACTCGCC-3′), the right-side-up filled triangles represent a single ribose at the cytosine (5′-AGGGArCTCGCC-3′), and the black diamonds are both positions with a ribose (5′-AGGGrArCTCGCC-3′). Single-turnover assays were conducted in triplicate at room temperature with 1.5 μm CcrM C. crescentus, 1 μm substrate and saturating cofactor AdoMet 15 μm; standard errors are shown.