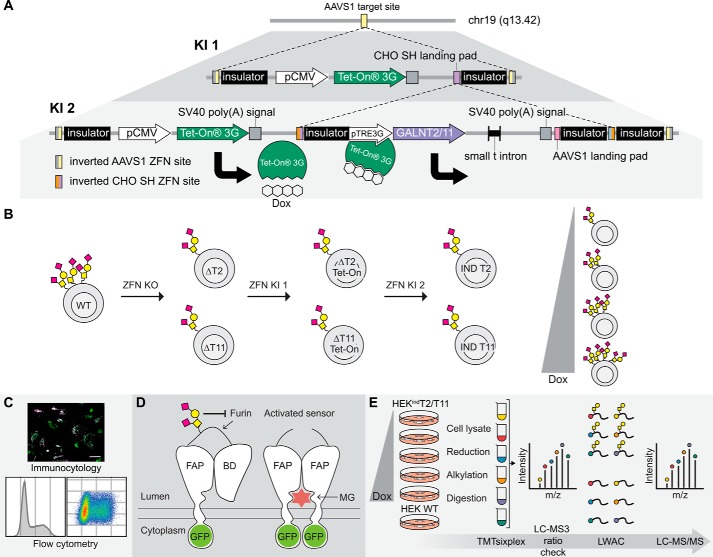
Figure 1.
Generation, validation, and exploration of isogenic HEK cell lines capable of doxycycline-inducible expression of GalNAc-T2 and -T11. A, overview of the tandem KI strategy. The first ZFN-mediated KI directs integration of the pCMV-TETOn3G cassette at the adeno-associated virus “safe harbor” (AAVS1) locus. A second KI targeted to the CHO SH landing pad within the first cassette stacks either GALNT2 or GALNT11 under control of the inducible pTRE3G promotor at the same locus. The inducer, doxycycline, binds to the Tet-On 3G protein, thereby enabling Tet-On 3G to complex with the pTRE3G promotor and drive transcription of the inducible gene. B, HEK ZFN knockouts of GALNT2 (ΔT2) and GALNT11 (ΔT11) were generated and subsequently subjected to the tandem KI strategy to generate HEKindT2 and HEKindT11 monoclonal cell lines. C, the inducible cell lines were validated by immunofluorescence, and HEKindT2 was validated also by flow cytometry. D, a GalNAc-T2 biosensor was used to validate activity of induced GalNAc-T2. The biosensor consists of a fluorogen-activating protein (FAP) and a blocking domain (BD) connected via a linker containing a GalNAc-T2–specific glycosylation site and a furin cleavage site. The sensor traffics via the Golgi en route to the cell membrane, and the absence of GalNAc-T2–mediated glycosylation leads to furin cleavage of the linker, departure of the blocking domain, and dimerization of the fluorogen-activating protein domains, which at the cell surface bind and activate the dye MG. The biosensor is GFP-tagged, allowing ratiometric measurement of activation. E, workflow for generating TMTsixplex-powered quantitative differential O-glycoproteomes for HEKindT2 and HEKindT11. Six parallel cultures, five HEKindT2 or HEKindT11 and one HEK WT, were cultured for 72 h, and the inducible cell lines were differentially induced for the final 48 h. Tryptic peptides from cell lysates of respective cultures were TMT-labeled, the six samples were combined, and a 1% aliquot was analyzed by LC-MS3 (ratio check). Subsequently, O-glycopeptides were enriched from the remaining sample by LWAC and analyzed by LC-MS/MS for O-glycopeptide identification and quantification. Finally, a deep proteome analysis was performed on the LWAC FT.