Figure 5.
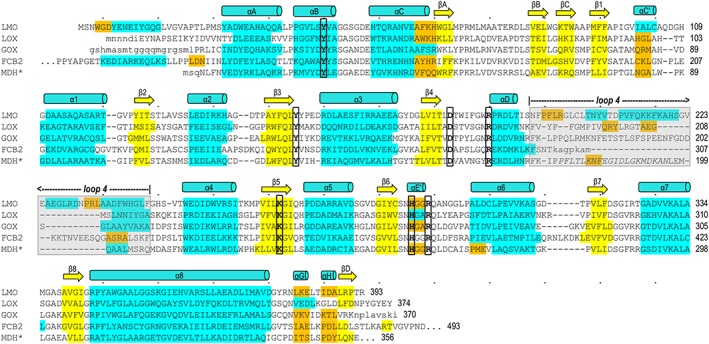
Structure‐based sequence alignment of LMO with representative α‐hydroxy acid oxidase family members. LMO (WT sequence, structure from C203A chain A) is shown first followed by LOX (PDB entry 2E77 chain B), GOX (2RDU chain A), FCB2 (1KBI chain A), and MDH* (MDH‐GOX chimera with chimeric region from spinach GOX italicized; 1P4C chain A). Conserved active site residues (bold, black outline) and residues in α‐helices (cyan), 310‐helices (orange), and β‐strands (yellow) are indicated. Secondary structural elements conserved among all family members are named in a manner consistent with naming conventions for other family members.6, 11, 39 Loop 4 (gray shading) is an active site lid that has been seen to undergo order/disorder transitions in other α‐hydroxy acid oxidases; when ordered, it covers the active site channel but is not structurally conserved between family members. Dots above the LMO sequence indicate every tenth character and a number is given for the residue at the end of each line. Residues in lower case letters are disordered in the respective structure.
