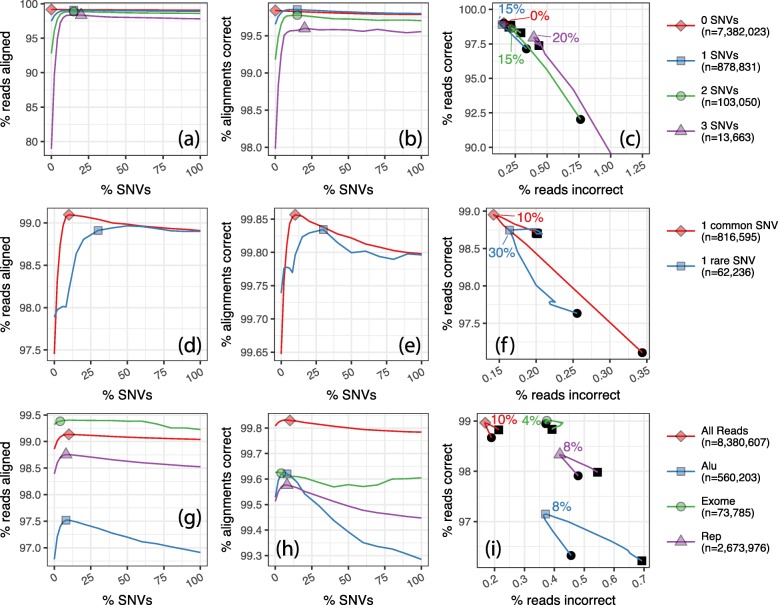
Fig. 3.
First row: Results for simulated reads stratified by the number of SNV alternate alternate alleles overlapped by the read. Reads overlapping regions with high DangerTrack [13] score—indicating the regions are difficult to align to—are omitted. Second row: Results for simulated reads overlapping exactly one common alternate allele (and no other alternate alleles) and reads overlapping exactly one rare allele. Reads overlapping regions with high DangerTrack [13] score, indicating the regions are difficult to align to. Third row: Results for simulated reads stratified by region of origin. Regions examined are regions labeled with the “Alu” family by RepeatMasker, regions captured by the Nextera exome sequencing protocol (“Exome”), and regions labeled with any repeat family by RepeatMasker (“Rep”)