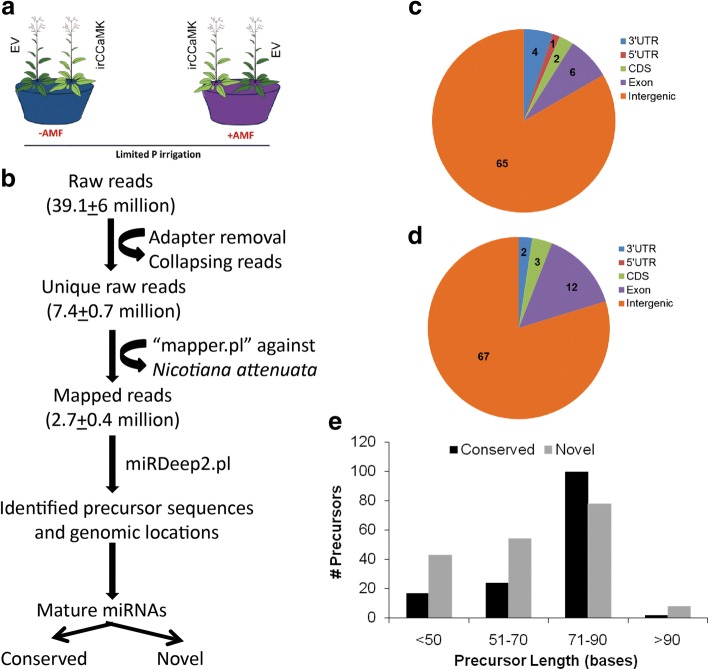
Fig. 1.
Elucidation of AMF-induced miRNA reprograming in Nicotiana attenuata roots. a Experimental set-up. Empty vector (EV) Nicotiana attenuata were competitively grown with an isogenic line silenced in the expression of a calcium and calmodulin protein kinase (irCCaMK) with and without out arbuscular mycorrhiza (+/-AMF) inoculum. Three replicate samples for each of the four conditions were used for library preparation and small RNA-sequencing. b Pipeline adapted to identify miRNAs in EV and irCCaMK plants with and without AMF inoculum. Counts for raw reads, unique reads, after removing adapter and collapsing, genome mapped reads, precursor sequences and genomic loci detected are provided at each step. All the filtering steps and their results are described in Material and Methods. c, d Genomic annotation for conserved and novel precursor miRNAs. Pie-charts show the distribution of precursor genomic loci of the Nicotiana attenuata genome for the conserved (c) and novel (d) miRNAs. The number of miRNAs originating from the 3’UTR, 5’UTR, coding sequences (CDS), exon and intergenic regions are shown. e Histograms show the size distribution of the precursor sequences for both, the conserved (black bars) and novel (grey) miRNAs. The x-axis represents the size of precursor sequences in bases and the y-axis shows number of unique precursor sequences