Abstract
Large-scale genomic studies have recently identified genetic variants causative for major neurodevelopmental disorders, such as intellectual disability and autism. However, determining how underlying developmental processes are affected by these mutations remains a significant challenge in the field. Zebrafish is an established model system in developmental neurogenetics that may be useful in uncovering the mechanisms of these mutations. Here we describe the use of voxel-intensity, deformation field, and volume-based morphometric techniques for the systematic and unbiased analysis of gene knockdown and environmental exposure-induced phenotypes in zebrafish. We first present a computational method for brain segmentation based on transgene expression patterns to create a comprehensive neuroanatomical map. This map allowed us to disclose statistically significant changes in brain microstructure and composition in neurodevelopmental models. We demonstrate the effectiveness of morphometric techniques in measuring changes in the relative size of neuroanatomical subdivisions in atoh7 morphant larvae and in identifying phenotypes in larvae treated with valproic acid, a chemical demonstrated to increase the risk of autism in humans. These tools enable rigorous evaluation of the effects of gene mutations and environmental exposures on neural development, providing an entry point for cellular and molecular analysis of basic developmental processes as well as neurodevelopmental and neurodegenerative disorders.
Keywords: neural development, voxel-based morphometry, deformation-field analysis, neuroanatomy, zebrafish
1. Introduction
Next generation sequencing methods and genome-wide association studies have revolutionized psychiatric genetics by identifying risk alleles for common neurological disorders. Applied to large patient cohorts, these methods have revealed numerous rare and de novo mutations, including copy number variants, small insertions or deletions, and single nucleotide polymorphisms, which cumulatively play a significant role in genetic risk for common neurodevelopmental disorders including intellectual disability, Tourette’s syndrome, attention deficit hyperactivity disorder and autism spectrum disorders (ASD) (reviewed in [1–5]). In addition to the strong genetic component associated with neurodevelopmental disorders, there is significant epidemiological data supporting environmentally-induced risk factors such as prenatal exposure to valproic acid [6].
Although key genetic and environmental risk factors have been identified, empirical evidence remains weak for specific risk-associated structural or functional changes within the brain. A major challenge now is to understand how these factors perturb neural development. The larval zebrafish is a model that enables a systematic search for subtle changes in brain architecture and function [7,8]. Live-imaging of the entire larval brain at single cell resolution allows visualization of whole brain microstructure and cell-type composition, and highly accurate registration of brains to a common reference facilitates comparisons between individuals. The zebrafish brain is built using molecular pathways conserved across vertebrates, and shares conspicuous neuroanatomical homologies with mammalian brains [9,10]. Moreover, zebrafish are an ideal system for analyzing the effects of gene mutations on brain development, because most human genes have a readily identifiable ortholog in zebrafish and mutants can be efficiently generated using Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR)-based genome editing [11,12]. Additionally, zebrafish are a cost-effective vertebrate system in which to perform in vivo chemical genetics experiments, allowing high-throughput evaluation of drug effects on the brains of wild-type and genetically mutant animals [13,14].
In humans, many genetic mutations strongly disrupt neural development, producing severe syndromic disorders present at birth. However, other neurodevelopmental disorders typically manifest only after several years of life, and are associated with less severe perturbations that are harder to detect [15–18]. Because it is difficult to systematically characterize subtle changes throughout the brain, many studies instead focus on candidate regions, potentially overlooking important anatomical or functional abnormalities elsewhere in the brain. The development of voxel-based morphometry (VBM) and its application to MRI studies in human has allowed whole brain analysis without a priori knowledge of regions that may be affected. VBM is widely used in human neuroimaging studies, allowing voxel-level comparisons of brains following registration to a common template, in order to identify statistically significant differences [19,20].
We and others have recently demonstrated that highly accurate brain registration can be achieved in larval zebrafish using non-linear registration methods [8,21–23]. Here, we capitalized on this ability to accurately register images by implementing brain morphometry in zebrafish, similar to that used in humans. Using simulated phenotypes, we estimate the sensitivity of voxel-intensity based methods for locating changes in neuronal composition. For identifying abnormalities in brain microstructure, we test the effectiveness of deformation-field analysis and atlas-based volume measurements. Statistical thresholds are calculated using permutation analysis within brain regions defined by a new computationally derived neuroanatomical map. This method discloses widespread neuronal changes in a zebrafish valproic acid model. We anticipate that the methods described here will enable systematic and stringent analysis of conserved neurodevelopmental processes as well as zebrafish models of human neurodevelopmental disorders, and that it will facilitate comparative work across model organisms.
2. Methods
2.1. Zebrafish husbandry
Zebrafish were maintained under standard laboratory conditions. Embryos were raised in E3 medium supplemented with 1.5 mM HEPES (pH 7.3) and 300 μM N-Phenylthiourea (PTU, Sigma P7629) starting at 8–22 hours post fertilization (hpf) to inhibit melanogenesis. Larvae were raised at 28°C on a 14h:10h light:dark cycle with medium changes every 2 days. Experiments were conducted in accordance with the National Institutes of Health guide for the care and use of laboratory animals and were approved by the National Institute of Child Health and Human Development animal care and use committee.
2.1.1. Zebrafish lines
Lines used in this study were Tüpfel long fin (TL); vglut2a:GFP (derived from TgBAC(slc17a6b:lox-DsRed-lox-GFP)nns14 by Cre injection) [24]); TgBAC(gad1b:lox-RFP-lox-GFP)nns26 (gad1b:RFP) [24]; Tg(evx2:Gal4)nns52, Tg(elavl3:ubci-Cer-sv40)y342 (huC:Cer) [21]; Tg(isl1:GFP)rw0, Tg(elavl3:lyn-tagRFP)mpn404 (huC:lyn-RFP), TgBAC(foxb1a:Gal4-vp16) [25]; Tg(elavl3:Gal4-vp16)nns6 (huC:Gal4) [26]; Tg(UAS-E1b:Synaptophysin-TagRFPT)y261 (UAS:syp-RFP) [27]; y271Et, Tg(actb2:loxP-eGFP-loxP-ly-TagRFPT)y272 (βactin:Switch) [28]; Tg(elavl3:Hsa.HIST2H2BE-RFP)a154 (huC:h2b-RFP) [8]; Tg(atoh7:GFP)rw021 [29]. Construction of Tg(−2.6Cau.tuba1:mCar.zf1)y516 (tuba:mCar) and Tg(elavl3:nls-mCar.zf1)y517 (huC:nls-mCar) will be described in detail elsewhere. Briefly, tuba:mCar uses the goldfish α-tubulin promoter, and huC:nls-mCar the zebrafish elavl3 promoter, to express the far-red fluorescent protein mCardinal, optimized for zebrafish [28,30,31].
2.1.2. Morpholino knockdown
For morpholino experiments, 4.6 nL of a 200 μM stock of a previously-established antisense morpholino (TTCATGCGTCTTCAAAAAAGTCTCC) was injected into huC:mCar transgenic larvae at the one cell stage to knock down atoh7 expression [32]. When injected into larvae with labeled retinal ganglion cells (RGCs), this dose is sufficient to suppress RGC pathfinding out of the retina and into the larval brain. In pilot experiments morpholino efficacy was confirmed through injection into Tg(atoh7:GFP)rw021 transgenic fish, which label retinal ganglion cells. As expected, injection prevented formation of the optic tract, blocked the behavioral response to a dark flash and resulted in hyperpigmentation, a marker of blindness in zebrafish larvae (Supplementary Figure 1).
2.1.3. Valproic acid treatment
Triple transgenic tuba:mCar, vglut2a:GFP, gad1b:RFP larvae were treated with 50 μM valproic acid (Sigma P4543) in E3 supplemented with PTU from 7 to 56 hpf and then maintained in E3/PTU until imaging at 6 dpf. We only show and discuss brain regions that were significant in two independent experiments, and for further stringency, focus on clusters of significant voxels that were approximately bilaterally symmetrical. Measurement of startle responsiveness to an acoustic stimulus was performed as previously described [33].
2.2. Image acquisition
Live 6 dpf larvae were anesthetized with 0.24 mg/mL tricaine methanesulfonate (MS-222), mounted in 2.5% low melting point agarose, and imaged using a Leica TCS SP5 II inverted laser-scanning confocal microscope [22]. Images were acquired using a 488 nm laser with spectral window set to 500–550 nm, a 561 nm laser with a spectral window of 566–624 nm, and a 633 nm laser with spectral window set to 638–795 nm. Dye separation was performed using Leica Application Suite Advanced Fluorescence software, with image volumes stitched and channels split using Fiji software prior to registration [34,35]. Brain scan resolution was isotropic 2 μm per voxel.
2.3. Neuroanatomical segmentation
2.3.1. Pre-processing
Input data for neuroanatomical segmentation were the brain-wide expression patterns for 210 transgenic lines [21,22]. For each line, brains of 3–10 larvae were live imaged, registered to a common reference and averaged to create a representative pattern of transgene expression. The resulting images were 8-bit grayscale with a spatial resolution of 1030 × 616 × 420 (nx × ny × nz) voxels (where 1 voxel = 1.0 μm). Volumes were masked to consider only those voxels within the brain. Thus, each voxel inside the brain had a gene expression vector consisting of the 210 transgene expression intensities at that point.
To ensure meaningful distance measures between voxels, we first normalized the set of 210 features assigned to each voxel so that all transgenic line images were scaled to have the same mean value of 127, among the voxels within the brain mask, with values outside the range [0,255] clipped after scaling. We tested alternate methods for normalization of gene expression, including manually determined thresholds, but none outperformed the simple scaling scheme above. Next gene expression images were median filtered at the original resolution using a spherical footprint within a 5 × 5 × 5 cube (median among total of 57 closest voxels). Our masked brain included ~ 42.2 M voxels. Because clustering that many voxels exceeded our computational capacity and time constraints, we down-sampled images by a factor of 2, assigning to each voxel the mean value within the initial corresponding 2 × 2 × 2 cube. Thus, our neuroanatomical maps (ROIs) were generated at half of the original resolution, i.e. 515 × 308 × 210 voxels, with the brain mask now comprising 5.25 M voxels.
2.3.2. Clustering
To derive neuroanatomic regions, we used k-means clustering to efficiently group all voxels into K clusters based on their Euclidean distances to K iteratively updated centroids. We explored maps with the range of the user-defined parameter K between 100 and 1000. We used the implementation of k-means in the python package sklearn, which we ran on NIH Linux cluster Biowulf (https://hpc.nih.gov/systems).
2.3.2.1. Addition of voxel geometric/spatial information
Clustering based on expression vectors without regard to spatial information treats voxels as independent samples and produced clusters that were frequently not contiguous in 3D space. Therefore, to include geometric information about voxel spatial relationships, we added voxel coordinates to the Ng=210 genetic features for a total of 213 features. For proper scaling, we multiplied each coordinate by a factor fs which weighs the relative importance of the spatial information. Values of fs that are too low produce non-contiguous regions, whereas high values tend to over divide large or elongated regions purely due to the large underlying spatial distance between constituent voxels. Therefore, the integer voxel coordinate values, x, y, and z, (starting with 0) were scaled according to:
| (1) |
where nmax=max(nx , ny , nz) before being added to the gene expression vector.
This means that for fs=10, the maximal distance between the voxel’s spatial features is comparable to the maximal distance among Ng gene expression features. In practice, much smaller values than that were needed, and we explored the range fs∈[0,2].However, we found that no single value was sufficient to directly produce an ideal neuroanatomical segmentation. When fs was large enough to prevent over fragmentation, larger and more elongated anatomical regions, such as structures within the optic tectum, were too subdivided. We therefore used intermediate values and introduced post-processing to resolve spatially non-contiguous clusters (2.3.3).
The larval zebrafish brain is largely symmetrical. To allow bilaterally symmetric regions to belong to the same cluster, we determined the plane of the bilateral symmetry (BS-plane) using our brain mask and assigned all coordinates on one side of the BS-plane to those of their bilaterally symmetric pair. In practice, since BS-plane was approximately parallel to the x-z plane, we simply adjusted the y coordinates according to yBS = (y - yc) where yc=155 defined the BS-plane in the image coordinates at half the original resolution.
2.3.2.2. Dimensional Reduction with LDA
Results obtained using the steps described above contained significant salt-and-pepper noise in cluster assignments and failed to discriminate some important neuroanatomical divisions. We reasoned these failures may in part have been due to the large number of features. To reduce the dimensionality of our feature space and improve discrimination of anatomical regions, we used linear discriminant analysis (LDA). This optimization method finds a set of linear combinations of the original features that maximizes the ratio of the between-class to the within-class variability,
| (2) |
where w is the vector of coefficients for the linear combination of features, and SB and Sw are the between-class and within-class covariance matrices, respectively. The solution is obtained by solving a generalized eigenvalue problem
| (3) |
which allows selection of the subspace with the NLDA that most discriminates features based on the magnitude of their eigenvalues. One needs a labeled set of voxels for which there is a known classification to obtain such a subspace. We used the Z-Brain regions, focusing on a subset of 73 subdivisions that represent neuroanatomical entities [8]. LDA dimensional reduction of our original 213 features (i.e. 210 expression patterns plus 3 spatial coordinates) derived a set with NLDA=75 features, that was subsequently used as input for k-means clustering.
2.3.3. Post-processing
Clustering results still contained small “islands” of disconnected ROIs for many clusters, as well as traces of salt-and-pepper noise in cluster assignments. To reduce salt-and-pepper noise, we applied median filtering with radius rm=1, or rm=2. In addition, we detected isolated islands by studying the network of voxels for each cluster and identified connected network components, again accounting for bilateral symmetry by treating the bilaterally symmetric voxel pairs as neighbors. For each cluster c, the largest contiguous sub-cluster was preserved as belonging to the same cluster c, while disconnected components were re-assigned to a neighboring cluster that surrounded them. If multiple clusters assignments were found among the immediate neighboring voxels that surrounded the “island”, its cluster index was assigned based on majority vote (the mode of the cluster index distribution). Ten iterations were sufficient to remove all disconnected components in most segmentations, and in the few remaining cases, where a fraction of voxels remained unassigned (usually < 10), they were removed and treated as background.
2.3.4. Validation
We explored the clustering algorithm by creating 1206 parameter sets containing different values for the presumed number of clusters K in k-means, weighting of genetic versus spatial information (fs), dimensional reduction schemes and post-processing methods. To select the map that best recapitulated known anatomical regions, we defined a benchmark set of 34 ROIs that were conservatively drawn within discrete, easily recognized brain regions. We then assessed each cluster-derived map by calculating its F1 score and Fowlkes-Mallows index compared to the benchmark regions [36]. We inspected maps that scored in the top 10 of either measure and selected a map with K=180 regions, that appeared in both top groups, and that also best segmented additional regions (e.g. Mauthner cell and torus longitudinalis) not used for validation. To estimate the symmetry of this map, we scanned voxels in one hemisphere and for each, explored the neighborhood of its bilaterally symmetric pair (within a 2 μm radius), checking whether any voxels had the same cluster assignment.
2.3.5. Segmentation of cellular/neuropil regions
To obtain masks that distinguished cellular and neuropil zones, we performed the same clustering procedure as described above, but this time using only the following six transgenic lines as features: βactin:Switch, huC:h2b-RFP, huC:lynRFP, huC:nls-mCar, huC:Gal4;UAS:syp-RFP, y271-Gal4. Additionally, in this case we used no geometric information (fs=0) and allowed for the existence of non-contiguous regions. Besides performing the clustering using such a 6-dimensional feature space, we also used LDA with equal or reduced number of dimensions (NLDA=2–6). The labeling for LDA was provided in the form of two classes, cell and neuropil regions that were obtained using smoothing and thresholding of huC:nls-mCar and huc:Gal4;UAS:syp-RFP images, to obtain a good initial guess for the cell region. Clustering was performed using K from 2 to 15. The optimal map was selected through manual comparison to the expression of huC:nls-mCar and huC:Gal4, UAS:syp-RFP patterns. In this map, with K=3, two clusters together covered neuron-rich regions, while the remaining cluster primarily contained neuropil but also comprised ventricular regions, glia and proliferative zones. This mask tends to assign regions where neuronal somas and neuropil are mixed to the cellular zone. Its primary deficiency is assigning too much of the pallial neuropil zone to the cellular cluster.
2.4. Brain morphometry
2.4.1. Image Registration
Image registration was performed using Advanced Normalization Tools (ANTs, RRID:SCR_004757) version 2.1 running on the National Institutes of Health’s Biowulf Linux computing cluster, using the parameter set optimized for live-imaging [22,37]. For valproic acid experiments, we performed multi-channel registration, equally weighting the tuba:mCar, vglut2a:GFP and gad1:RFP images.
2.4.2. Smoothing and normalization
Following registration and prior to statistical analyses, images were smoothed with a 3-dimensional boxcar average using a width of a cell radius (diameter ~8 μm) to remove subcellular details and to reduce inter-subject variability. Smoothing results in each voxel representing a weighted mean of its own and neighboring voxels’ values, reducing the influence of imaging noise. The voxel intensity range was then linearly normalized to [0 to 1000] to account for differences between brain scans, saturating the top and bottom 0.1% of pixel intensities to minimize scaling distortion due to outlier values. Normalization was not performed for deformation field images.
2.4.3. Voxel-intensity comparisons
Voxel-wise comparisons were performed only on pixels within a mask encompassing the brain, excluding non-neural tissue and background. Rather than evaluate normality of the intensity distribution for each voxel, we first rank-transformed values at each voxel before performing t-tests between groups. For voxel-wise comparisons across the whole brain (5.3 M voxels), we controlled the family-wise error rate for α=0.05 using the Holm-Bonferroni procedure.
2.4.3.1. Permutation-testing for region-wise thresholds
We performed 1000 permutation tests to establish a non-parametric significance threshold for each neuroanatomical region. In each permutation trial, we randomly assigned brain images to two groups, while maintaining the original sample size of each group. We then calculated t-tests as above across all voxels in the brain. For each permutation, we recorded the maximal and minimal t-statistic values for each region. After 1000 such trials, we then ranked all the maximal t values for each region, and took the 990th ranked score for each region as representing the threshold that was attained by only 1% of random trials. Thus, all voxels within a region where the t-statistic between actual comparison groups exceeded the threshold for that region were considered significant at α=0.01. We also ranked minimum t-statistic values to identify voxels with significant intensity differences in the opposite direction.
2.4.3.2. Analysis of clusters of nominally significant voxels
To find clusters of nominally significant pixels, we first built a brain-wide map of t-statistics (2.4.3), then formed clusters of contiguous voxels with t-statistics corresponding to unadjusted significance values of p < 0.0001. For each cluster, we calculated a metric (Tcluster) which was the sum of the absolute values of the t-statistics of all constituent voxels. This measure was chosen as it can represent both large clusters of weakly significant pixels and small clusters of strongly significant pixels. We used permutation analysis to establish significance thresholds for clusters: we randomly assigned images to two groups, calculated t-statistics for all voxels, performed cluster analysis, and recorded the most positive and most negative Tcluster values. This process was repeated 1000 times to find the Tcluster value exceeded by only 5% of randomly assigned groups. This value was then used as the significance threshold in comparisons between the original groups.
2.4.4. Deformation field analysis
The non-linear diffeomorphic transformation yields a deformation field that describes how each voxel was adjusted to match an individual experimental brain to the reference brain. The change in volume of each voxel can be derived from the determinant of the Jacobian matrix. Taking the log of the determinant results in a map in which negative values indicate decreases in voxel volume of the registered image and positive values indicate increases. We used the ANTs CreateJacobianDeterminantImage function to generate a map of the log of the Jacobian determinant. Voxel- and cluster-wise statistical analyses were applied as described above (2.4.3).
2.4.5. Atlas volume measurement
To measure the volume of the whole brain and subdivisions, we used the ANTS antsApplyTransforms command (MultiLabel interpolation) and the InverseWarp transformation matrix produced during diffeomorphic registration, to back transform the neuroanatomical and cell/neuropil maps (described in Methods section 2.3) to the original unregistered brain images. We then measured the size of each brain (total pixel count across all regions), cell-rich and neuropil volumes and the volume of each neuroanatomical region, and applied t-tests with Holm’s correction to compare volumes between experimental groups.
2.5. Image processing for simulated brain abnormalities
For simulating misplaced cells within specific brain regions, we scanned brains from 40 vglut2a:GFP larvae. Images were registered to the vglut2a:DsRed reference brain using ANTs. We then used the ANTs antsApplyTransforms command with the InverseWarp transformation matrix to back-transform our neuroanatomical region map to each original image stack, using MultiLabel interpolation to preserve the categorical region indices. This enabled us to locate the ventral thalamus, posterior tuberculum and inferior olive in original brain scans. We then randomly placed 7.6 μm diameter ‘cells’ within each selected region, altering a total of 26 voxels for each cell. Addition of 300 cells thus changed around 17% of voxels in the ventral thalamus (total 46672 voxels in our mask). The intensity at the center of the cell was set to the maximal image intensity within the selected region with decreasing intensity toward the edges. Finally, we registered the processed image stacks to the reference brain. The processed and original images were both present in the dataset that we used when randomly sampling to form comparison groups, so in some cases, groups may have included processed and unmodified versions of the same brain. For inverting brain scans around the AP axis, we used ImageJ [38].
2.6. Statistics and software
Data in figures and text show means and standard errors. In all panels, unless otherwise noted: horizontal and sagittal views are displayed with anterior to the left, coronal views with dorsal side up. CobraZ software for morphometric analysis was written in IDL, runs under the IDL virtual machine (freely available from www.harrisgeospatial.com) and the desktop version can be downloaded from our website (https://science.nichd.nih.gov/confluence/display/burgess/Software). IDL code and bash scripts for running in a linux cluster environment are available on request.
3. Results
Many neurodevelopmental disorders include a localized change in the density of specific neuronal cell types, arising from changes in neuronal proliferation, migration or differentiation, and may also involve subtle changes in the size and/or shape of specific brain regions. To perform an unbiased brain-wide analysis that is able to identify such changes, we implemented procedures for voxel-based morphometry, similar to methods that are widely used to statistically compare brain scans from normal individuals and patient groups. With this objective, we developed three methods for brain morphometry in zebrafish. The first method utilizes two measures for comparing voxel-intensity between groups: region-wise permutation analysis and cluster-analysis (sections 3.1-3.3). The second method identifies local changes in brain shape based on deformation field analysis (section 3.4). The third method also detects changes in brain shape, but measures volumes of brain subdivisions (section 3.5). The sections below describe these procedures.
3.1. Identifying local changes in neuronal density using voxel-intensity morphometry
To test the feasibility of voxel-intensity based morphometry in zebrafish, we acquired 40 whole-brain confocal images of transgenic vglut2a:GFP zebrafish that express green fluorescent protein in glutamatergic neurons. To simulate a neurodevelopmental abnormality affecting local cell density, we added 300 randomly positioned ‘cells’ to the ventral thalamus in each image volume (Figure 1A). We then registered unprocessed and modified images to a reference brain, so that corresponding voxels in each image were spatially aligned. Next we tested for differences in mean intensity between the original and processed image sets at every voxel using t-tests. To control the family-wise error rate over the large number of comparisons (5.3 M pixels), the Holm-Bonferroni method was used to identify statistically significant changes in pixel intensity (see Methods 2.4.3).
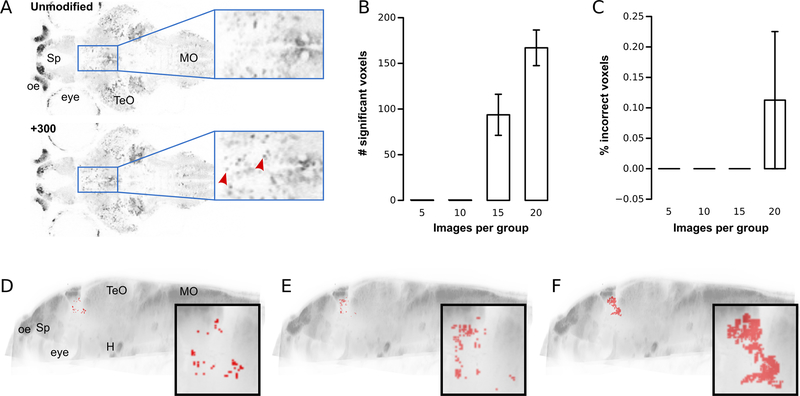
Figure 1. Voxel-intensity morphometry: sensitivity and error-rate.
A.Horizontal confocal slice from vglut2a:GFP transgenic zebrafish. Top panel: unmodified image. Bottom panel: similar level slice from a different brain scan, with 300 simulated cells added to the thalamus. Boxed regions show magnified views. Arrowheads mark simulated cells.
B.Number of statistically significant voxels in the thalamus after Holm-Bonferroni correction, using different numbers of brain scans per comparison group.
C.Percent of significant voxels located outside the thalamus for different size comparison groups.
D-F. Sagittal brain projection, showing location of pixels for comparisons between unprocessed and modified brainimages (15 images per group) that remained significant after Holm-Bonferroni correction (D), region-wise permutation testing(E), and cluster analysis (F). Insets show magnified views of the ventral thalamus.
H: hypothalamus; oe: olfactory epithelium; Sp:subpallium; MO:medulla oblongata; TeO:optic tectum.
To validate this procedure, we randomly selected 15 unprocessed and 15 modified images from the whole set, computing (1) the number of statistically significant voxels within the ventral thalamus, and (2) the false-positive rate (% of significant voxels outside the ventral thalamus). We performed this procedure 6 times, randomly sampling a different set of 15 brains per group each time, to calculate the mean sensitivity and false-positive rate. This method detected almost 100 voxels with greater mean intensity in the ventral thalamus (representing 0.2% of its total volume), with no false-positives elsewhere in the brain (Figure 1B-D). We then assessed the sensitivity of the procedure to the number of brain scans employed by using 5, 10 and 20 images per group (again, performing 6 repeats for each sample size). No differences were detected when we used fewer than 15 scans per group, whereas we identified almost twice as many significant voxels when using 20 scans per group (Figure 1B).
Although the Holm-Bonferroni correction produced a very low false positive rate, it has previously been shown that similar methods for controlling the error rate of parametric statistical tests for voxel-based brain comparisons are highly conservative [39]. Instead, empirically determining a suitable significance threshold for a given data-set using permutation tests has been shown to rigorously control false-positives while increasing sensitivity. For permutation testing, images are randomly assigned to two groups, t-tests performed for every voxel, and the most statistically significant p-value recorded. This procedure is repeated 1000 times. The resulting p-values are then ranked and the 50th ranked p-value used as a threshold for identifying differences between the original image groups that are significant at p < 0.05. However, in our tests, whole-brain permutation analysis did not identify more voxels in our simulated data set than when controlling error using the Holm-Bonferroni method (not shown). This is likely because substantial differences in variability between brain regions reduces the sensitivity of whole-brain permutation to small changes in regions with relatively low normal variability. We therefore decided to use permutation testing to separately establish significance thresholds for individual brain regions. To enable such region-wise comparisons, however, we first needed a detailed neuroanatomical map of the brain.
3.2. Identifying local changes in neuronal density using region-based morphometry
3.2.1. Automated segmentation of the larval brain
Neuroanatomical annotation typically uses histological stains to guide expert segmentation of anatomical features. The Z-Brain atlas describes such a set of larval zebrafish brain regions derived through manual segmentation [8]. We recently transformed these region masks to the Zebrafish Brain Browser reference brain that we use to register live brain scans [22]. However, manual labeling is an inherently subjective process that is prone to persistent biases and errors, often driven by prior misconceptions and difficult to perform comprehensively. Annotation is particularly difficult in the larval zebrafish brain, where neuroanatomical knowledge is limited and few regions have conspicuous nuclear organization so that boundaries between many structures are poorly differentiated.
Previous work in several species has demonstrated that gene expression data can be used to computationally derive anatomically accurate brain maps [40–42]. Having 3D images of transgene expression available from more than 200 zebrafish lines, we reasoned that a common pattern of gene expression among neighboring voxels may provide an accurate and unbiased indicator of the underlying anatomical structure. Consequently, we developed a computational method to create a neuroanatomical map of the 6 dpf larval zebrafish brain based on co-registered three dimensional transgene expression data (see Methods 2.3). To reduce the feature space, we applied linear discriminant analysis based on 73 manually segmented brain regions from Z-Brain. The essential part of our methodology is the clustering of image voxels based on their individual pattern of gene expression levels. We imposed a constraint that all voxels belonging to the same cluster form a contiguous region in three-dimensional space. Additionally, we account for bilateral symmetry of the brain, by relaxing this constraint, so that paired voxels on opposing hemispheres are allowed to belong to the same cluster.
We evaluated the results of different clustering methods and parameters by comparing computationally identified regions to a set of 34 conservatively outlined neuroanatomical regions that were identified by a human expert (see Methods 2.3.4). The top ranking map derived by this procedure comprised 180 regions (Figure 2A-C, Supplementary File 1). Many of these regions are easily recognized, and similar to manually segmented structures in Z-Brain (Fig. 2D). In other cases, multiple regions in the computational map additively correspond to a single region in Z-Brain, possibly revealing anatomical subdivisions that are difficult to manually segment or that were previously unrecognized (Fig. 2E). In the hindbrain, which is particularly difficult to annotate, some computational regions align well with longitudinal stripes of transcription factors that are thought to be an organizational principle in this area (Fig. 2F)[43]. The zebrafish brain is bilaterally symmetric with the exception of the dorsal habenula. Importantly, our clustering algorithm allows voxels on opposite sides of the axis of symmetry to belong to the same cluster, but does not impose bilateral symmetry. This means that pairs of regions on the left and right hemispheres can have different shapes. Nevertheless 92% of interhemispherically paired voxels were assigned to the same cluster, with most unmatched voxels at cluster boundaries (not shown), confirming that the map substantially captured intrinsic symmetry. Thus, the computational segmentation method reveals known neuroanatomical regions, and may disclose previously unrecognized structure within the larval brain.
Figure 2. Computationally derived neuroanatomical segmentation.
A.Sagittal brain slice showing computationally derived regions and planes corresponding to the sections in B and C. The 3D neuroanatomical atlas is included within the CobraZ package, available at https://science.nichd.nih.gov/confluence/display/burgess/Software.
B.Coronal slices showing regions through the forebrain (1), midbrain (2), cerebellum (3) and hindbrain (4)
C.Horizontal slices showing regions through the tectum (1), tegmentum (2) and hypothalamus (3).
D.Comparison of computational regions (black outlines) to manually segmented anatomical regions in Z-brain (colored regions). Horizontal views (top panels, anterior up) and coronal views (bottom panels) are shown. Left to right: habenula, dorsal thalamus, nucleus of the median longitudinal fasciculus, hypothalamus, inferior olive.
E.Comparison of computational regions (colored regions) to Z-brain regions (outlines). Left to right: horizontal views of optic tectum neuropil, pallium and cerebellum.
F.Top panel: computational regions in the hindbrain. Bottom panel, same regions outlined, showing transgenic expression of evx2:Gal4 (magenta), isl1:GFP (blue) and foxb1a:Gal4 (green).
TeO: optic tectum; H:hypothalamus; MO:medulla oblongata; Ce:cerebellum; Sp:subpallium; Pa:pallium; Hb:habenula; Th:thalamus; nMLF:nucleus of the medial longitudinal fasciculus; IO:inferior olive
3.2.2. Region-based permutation analysis of differences in voxel intensity
Our comprehensive map of larval brain regions enabled us to use permutation tests to separately establish significance thresholds for each of the 180 regions (see Methods 2.4.3.1). We compared the sensitivity and false-positive rates of these thresholds to those established using Holm-Bonferroni correction, using the simulated data set with 300 additional ‘cells’ described above. Region-wise thresholds correctly identified around 50% more voxels within the ventral thalamus, both for N=15 and N=20 images per group (Figure 1E, 3A). False positives rates remained low, representing fewer than 0.1% of all voxels.
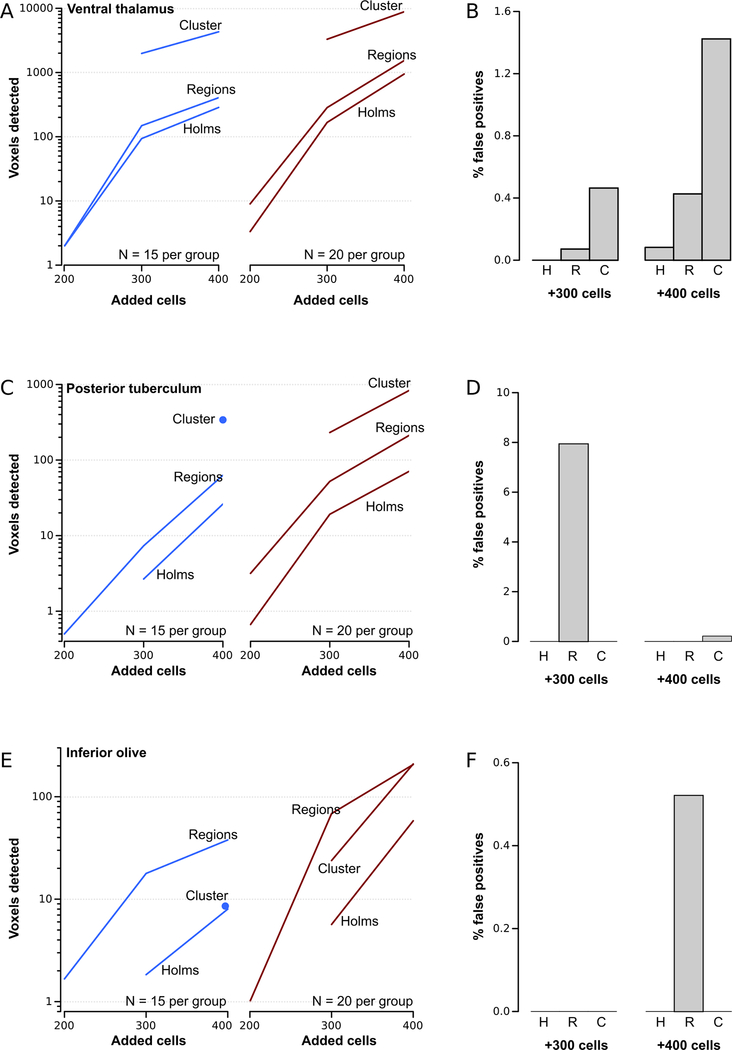
Figure 3. Region-wise permutation testing and cluster-analysis of voxel-intensity differences.
A.Number of significant voxels detected in the ventral thalamus for N=15 brains per group (left graph) and N=20 brains (right graph) for different numbers of simulated cells added to the ventral thalamus. Significant pixels were identified using Holm-Bonferroni correction, region-wise permutation analysis and cluster analysis as indicated.
B.Percent of significant pixels detected outside the area where simulated cells were added, when significance thresholds were established using Holm-Bonferroni correction (H), region-wise permutations (R) and cluster analysis (C).
C-F As for A and B, but where simulated cells were added to the posterior tuberculum (C-D) or the inferior olive (E-F).
To more thoroughly compare methods for establishing significance thresholds, we used our image processing step to introduce 200 or 400 ‘cells’ into the ventral thalamus, then repeated the comparisons. When only 200 misplaced cells were introduced, neither method was sufficiently sensitive to correctly locate more than 10 significant voxels. With 400 cells, region-wise thresholds continued to correctly identify approximately 50% more significant voxels than Holm-Bonferroni thresholds. Indeed, using 20 brain scans per group, region-wise thresholds located more than 1500 voxels with greater mean intensity in processed images. False-positive rates remained low throughout.
Because the ventral thalamus has a low density of glutamatergic neurons, contrast introduced by our added neurons is high in this area. To test whether our procedure could identify additional glutamatergic neurons in regions of the brain with a greater density of glutamatergic neurons, we digitally added cells to the posterior tuberculum, which has a medium density of glutamatergic neurons and to the inferior olive which has a high density of glutamatergic neurons, and repeated voxel-intensity comparisons. For both brain regions, and across sample-sizes and number of added cells added, region-wise thresholds correctly identified a greater number of voxels (Figure 3C,E). As anticipated, voxel-intensity morphometry was less sensitive overall in these two brain regions than in the ventral thalamus. Region-wise thresholds were generally accurate, except where very few voxels were detected overall, so that the presence of a small number outside the correct region resulted in up to an 8% false positive rate (Figure 3D,F).
3.3. Analysis of clusters of voxels with nominally significant changes in intensity
Some changes may produce only sub-threshold changes at individual voxels within a large spatially contiguous area. We therefore searched for clusters of adjacent voxels, each with only a nominally significant mean intensity change (see Methods 2.4.3.2 ). [19]. Next, to integrate information the spatial extent of each cluster and the significance level of changes in its constituent voxels, we summed the t-statistic for all voxels within a cluster (Tcluster). Recent work has shown that spatial autocorrelation renders cluster-wise tests especially susceptible to false positives, and that the error-rate is best controlled by using permutation testing [39]. We therefore used whole-brain permutation testing to derive significance thresholds for Tcluster scores.
We tested this procedure using our dataset of brain images with added simulated cells. For the ventral thalamus and posterior tuberculum, which have a relatively low normal density of glutamatergic neurons, cluster analysis greatly increased sensitivity, accurately detecting up to 10-fold more voxels in the correct regions over voxel-intensity morphometry (Figure 1F, 3A,C). However, at the lowest added cell number used (200 extra neurons) cluster analysis failed to detect changes, possibly because there were too few contiguous voxels with mean intensity differences to form clusters. Likely for a similar reason, cluster analysis was less sensitive than voxel-intensity morphometry in the inferior olive, where simulated cells were added to a region with an already high density of glutamatergic neurons (Figure 3E,F).
3.4. Identifying changes in brain shape using deformation field analysis
Neurodevelopmental phenotypes may affect brain shape or size, for example by changing the relative volume of white and grey matter due to disruptions in synaptogenesis or cell proliferation [44–46]. However, non-linear registration algorithms may obscure such differences by enforcing shape-matching to the reference brain. This difficulty has been addressed by applying deformation field analysis to the transformation matrix that is created for each image during registration (Figure 4A)[47]. The Jacobian determinant of each point within the transformation matrix indicates whether the volume of a voxel in the original image was increased or decreased in order to match the corresponding voxel in the reference image. We therefore established a procedure to statistically compare the Jacobian determinants for two groups of brains (see Methods 2.4.4).
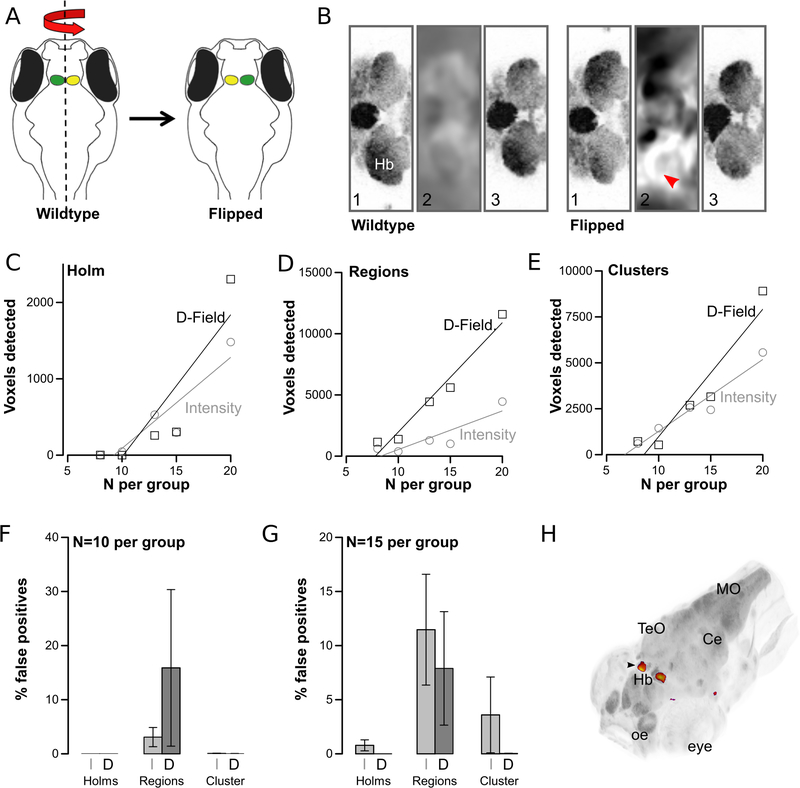
Figure 4. Deformation field analysis.
A.Processing of wildtype brain images by reflection around the anterior-posterior axis
B.Horizontal slice through the habenula showing (1) unregistered image, (2) deformation field produced during registration, (3) registered image. Left panels (wildtype) started with unprocessed images, right panels (flipped) with left/right-flipped images. Arrowhead shows voxel-expansion required on left side of image required to match reference brain.
C-E. Number of statistically significant voxels detected in the habenula after comparison of unprocessed and flip-brain images using Holm-Bonferroni correction (C), region-wise permutation testing (D) and cluster analysis (E). Results derived from analysis using voxel-intensity comparisons (grey circles) and deformation-field analysis (black squares)
F-G. False positives (percent of all significant voxels that were outside the habenula) for the experiment in C-E for N=10 (F) and N=15 (G) per group and using voxel-intensity analysis (I) or deformation-field analysis (J).
H. 3D-projection of significant pixels comparing normal and flipped brains, from region-wise permutations with N=15 per group. Arrowhead: habenula
Hb:habenula; MO:medulla oblongata; oe:olfactory epithelium; TeO:optic tectum; Ce:cerebellum
To test this method, we simulated a mutant phenotype, taking advantage of the inherent asymmetry of the habenula. The habenula consists of bilateral nuclei located in the dorsal diencephalon and is the only morphologically assymetric region of the zebrafish brain, with the left habenular nucleus larger than the right [48]. We scanned 40 wild-type brains with pan-neuronal expression of the far-red fluorescent reporter mCardinal and then reflected half of the scans along the anterior-posterior axis, so that the left and right sides were reversed (Figure 4A-B). We reasoned that a robust method for brain morphometry should identify changes only in the habenula in normal versus flipped brains.
After registration, we generated the log Jacobian determinant (LJD) image from the transformation matrix for each brain. We then compared deformation fields by applying voxel-intensity comparisons to the groups of normal and flip-brain LJD images. To test sensitivity, we selected subgroups of the images (8–20 per group), and repeated each analysis 6 times with different random selections. For comparison, we also performed voxel-intensity analysis on the registered brain images. Both voxel-intensity and deformation field analysis correctly located differences in the habenula. However, deformation field analysis tended to identify more changes (Figure 4C-E). For instance, with 20 images per group, cluster analysis of deformation fields identified 60% more voxels with significant differences in the habenula than voxel-intensity morphometry, with a false-positive rate of just 1.3% (Figure 4F-H). Overall, deformation field analysis using thresholds set by region-wise permutations was the most sensitive, but also showed a relatively high false positive rate of around 10%. Using either method we were able to correctly identify differences in the habenula using as few as 8 samples per group, establishing that local perturbations in brain shape can be reliably detected.
3.5. Identifying brain shape changes using atlas volume measurements
Our brain registration procedure is initialized with an affine alignment, including global scaling of each image to best match the reference brain. Therefore, voxel-intensity and deformation-field analyses may fail to detect changes that affect brain volume as a whole. We reasoned this could be detected by applying the transformation matrix produced during registration to the neuroanatomical map, in reverse, to align it to the original brain scan (see Methods 2.4.5). This enabled us to compute brain size, simply by measuring the volume of the whole back-transformed atlas, and similarly, record the original size of each brain region (Fig. 5A). We also used a similar approach to disclose changes that broadly alter brain cellular or neuropil sub-volumes. The set of 210 transgene expression patterns that we used for clustering included several that globally mark cell somas or neuropil. Therefore, to obtain a map of cell and neuropil regions, we repeated the clustering procedure using only those lines. The result was a map of brain regions that are strongly enriched for cell bodies or neuropil (Fig. 5B, Supplementary File 1). After registration, we could then transform this map back to original images and thereby estimate the total size of cell and neuropil dense regions.
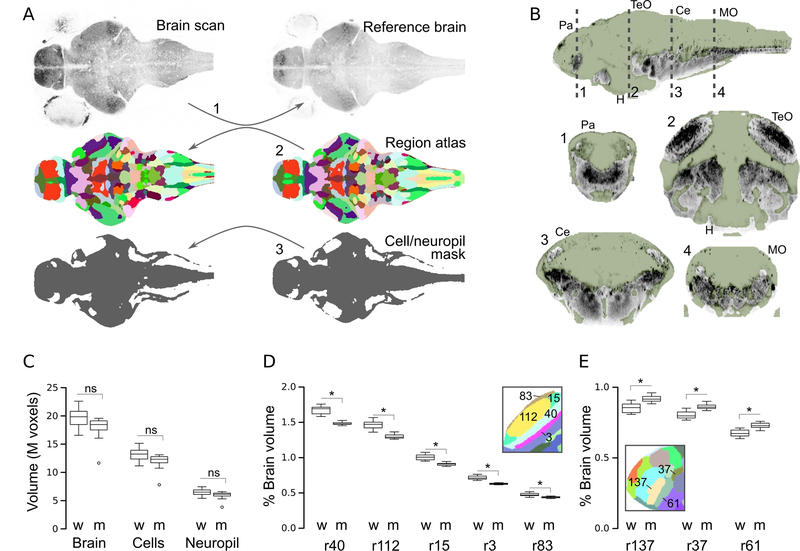
Figure 5. Atlas-volume measurements.
A.Schematic of procedure for measuring the volume of the whole brain, anatomical subdivisions and the cellular/neuropil zones. (1) Brain scans are registered to a reference. Then, the anatomical atlas (2) and map of cellular/neuropil regions (3), both generated on the reference brain, are back-transformed to the original images.
B.Cluster-derived map of cell-rich regions (green mask) and other (primarily neuropil, grey) brain regions. Dashed lines in top sagittal section indicate planes of section for corresponding coronal views below.
C. Volume of the whole brain, cellular regions and synaptic regions in wildtype larvae (w) and atoh7 morphants (m). N=11 per group.
D-E. Percent of brain occupied by regions (r) of the optic tectum neuropil (D) and forebrain (E) in wildtype (w) and atoh7 morphants (m). * p < 0.05 after Holm correction for 183 comparisons (ie.180 regions, whole brain, cell volume, neuropil volume). Insets show neuroanatomical map in the corresponding areas with the region numbers indicated.
MO:medulla oblongata; TeO:optic tectum; H:hypothalamus; Ce:cerebellum; Pa: pallium
To validate these measures, we generated embryos with morpholino knockdown of atoh7, an essential gene for optic tract formation, reasoning that loss of the optic nerve would reduce the size of the neuropil zone of the optic tectum. In contrast to atoh7 mutants that lack all retinal ganglion cells the transient activity of the anti-sense atoh7 morpholino only blocks differentiation of early-born retinal ganglion cells necessary for axon pathfinding out of the retina [32,49]. Brain volume, and the total volume of cellular and neuropil regions were similar in morphants (Fig. 5C). In our neuroanatomical map, tectal neuropil is subdivided into five regions, all of which were significantly reduced in size by around 12% in morphants; three subdivisions were top-ranked as different by both p-value and the magnitude of the change (Fig. 5D; Table 1). Two other retinorecipient nuclei, AF9 and the griseum tectale showed a similar reduction in size. Optic tectum cellular layers were also reduced in volume, potentially reflecting adaptation to decreased retinal input. Unexpectedly we also noted that morphants showed an increased size in adjacent regions in the forebrain, including the pallium and anterior commissure (Fig. 5E), in the interpeduncular nucleus and in two monoaminergic nuclei, the dorsal raphe and the locus coeruleus (Table 1). Atlas volume measurement is therefore a sensitive method for detecting changes that alter the size of specific brain regions.
Table 1.
Brain regions with significant volume changes in atoh7 morphants
| # | Region | wt | atoh7- | p-val |
|---|---|---|---|---|
| 137 | Anterior commissure | 0.853 ± 0.011 | 0.919 ± 0.007 | 0.018 |
| 116 | Griseum tectale | 0.688 ± 0.007 | 0.628 ± 0.006 | 0.000 |
| 033 | Interpeduncular nucleus tegmentum | 0.333 ± 0.003 | 0.361 ± 0.004 | 0.010 |
| 071 | Locus coeruleus | 0.224 ± 0.001 | 0.235 ± 0.002 | 0.026 |
| 069 | Medial tectal band | 1.626 ± 0.014 | 1.492 ± 0.013 | 0.000 |
| 086 | Mesencephalon | 0.451 ± 0.003 | 0.483 ± 0.005 | 0.010 |
| 042 | Migrated pretectal area M1 | 0.432 ± 0.004 | 0.406 ± 0.004 | 0.036 |
| 024 | Neuropil adjacent to IPN | 0.438 ± 0.003 | 0.474 ± 0.007 | 0.017 |
| 003 | Optic tectum - neuropil | 0.717 ± 0.008 | 0.627 ± 0.005 | 0.000 |
| 040 | Optic tectum - neuropil | 1.669 ± 0.019 | 1.481 ± 0.009 | 0.000 |
| 112 | Optic tectum - neuropil | 1.464 ± 0.02 | 1.303 ± 0.011 | 0.000 |
| 015 | Optic tectum - neuropil | 1.006 ± 0.013 | 0.908 ± 0.007 | 0.000 |
| 083 | Optic tectum - neuropil | 0.477 ± 0.007 | 0.439 ± 0.004 | 0.017 |
| 089 | Optic tectum - stratum periventriculare | 2.518 ± 0.033 | 2.218 ± 0.028 | 0.000 |
| 155 | Optic tectum - stratum periventriculare | 2.438 ± 0.027 | 2.204 ± 0.021 | 0.000 |
| 022 | Optic tectum - stratum periventriculare | 1.024 ± 0.011 | 0.939 ± 0.008 | 0.001 |
| 178 | Optic tract - AF9 | 0.335 ± 0.003 | 0.309 ± 0.003 | 0.001 |
| 037 | Pallium | 0.805 ± 0.009 | 0.863 ± 0.006 | 0.005 |
| 061 | Preoptic area | 0.675 ± 0.008 | 0.729 ± 0.006 | 0.006 |
| 162 | Rhombomere 1,2 - Ventral | 0.295 ± 0.003 | 0.326 ± 0.006 | 0.016 |
| 140 | Rhombomere 1,2 - Ventral | 0.28 ± 0.002 | 0.301 ± 0.004 | 0.035 |
| 080 | Rhombomere 7 | 0.42 ± 0.005 | 0.456 ± 0.006 | 0.035 |
| 060 | Rhombomere 7 - Dorsocaudal | 0.215 ± 0.002 | 0.23 ± 0.002 | 0.018 |
| 007 | Rhombomere 7 - Ventromedial | 0.484 ± 0.006 | 0.525 ± 0.007 | 0.043 |
| 079 | Superior raphe | 0.452 ± 0.003 | 0.482 ± 0.005 | 0.012 |
| 139 | Tegmentum | 0.857 ± 0.006 | 0.799 ± 0.01 | 0.011 |
| 147 | Tegmentum | 0.429 ± 0.003 | 0.455 ± 0.004 | 0.024 |
| 125 | Tegmentum | 1.202 ± 0.009 | 1.139 ± 0.01 | 0.026 |
3.6. Neurodevelopmental changes after valproic acid treatment
To test whether our voxel-intensity, deformation field and atlas volume methods could provide new insights into neurodevelopmental abnormalities, we used the valproic acid ASD model. VPA is a short chain fatty acid that is primarily used in humans as an antiepileptic and mood stabilizing drug. However, exposure to VPA during the first trimester of pregnancy has been linked to birth defects that include neural tube defects and congenital malformations. In addition, there is substantial evidence supporting an association between in utero exposure to VPA and an increased risk of developing ASD [6,50,51]. One theory is that autism is associated with a defect in the balance of excitatory/inhibitory signaling [52–55]. Therefore, in addition to assessing whole brain microstructure using pan-neuronal mCardinal expression, we examined effects on glutamatergic and GABAergic neuron composition using the vglut2a:GFP and gad1b:RFP lines respectively. Triple transgenic embryos were exposed to 50 μM valproic acid for 49 h starting at 7 hpf, a dose that we confirmed did not produce overt changes in development or behavior [56].
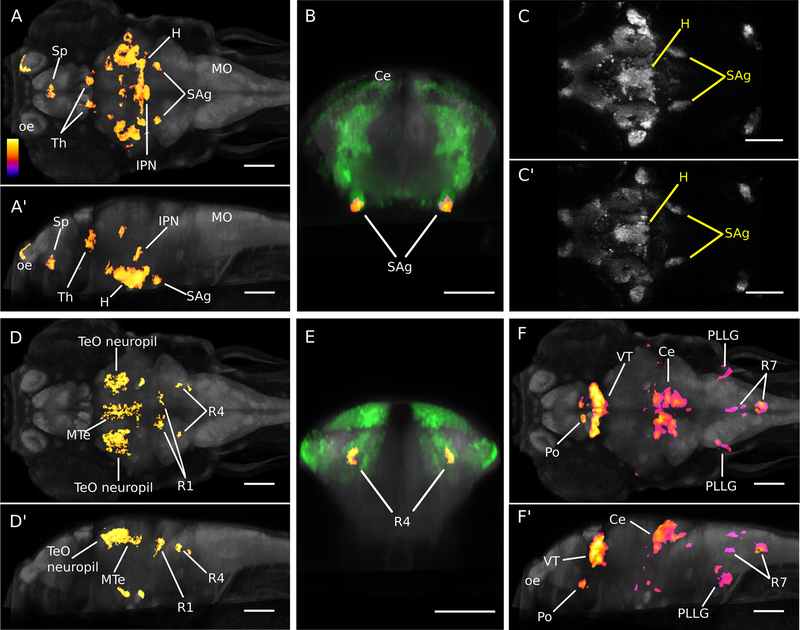
Treatment of larvae with VPA resulted in significant changes in both excitatory and inhibitory neuron composition as well as local changes in brain shape. Increased vglut2a:GFP signal was found only in the optic tectum neuropil, whereas reductions were observed in multiple brain regions including the subpallium, hypothalamus, thalamus, interpeduncular nucleus, and in an area at the anterior-most tip of the statoacoustic ganglion (Figure 6A-B). Visual comparison of matched slices from the original confocal images confirmed significant findings (Figure 6C). GABAergic neurons were also affected: treatment resulted in increased gad1b:RFP signal in the olfactory epithelium, olfactory bulb, and anterior commissure (data not shown) and a decrease in the optic tectum neuropil, medial tectal band, and rhombomeres 1 and 4 (Figure 6D-E). Deformation-field analysis revealed expansions of the preoptic area, thalamus, hypothalamus, cerebellum, the posterior lateral line ganglion and parts of rhombomere 7 (Figure 6F). The largest increases were seen in the cerebellum and ventral thalamus, where 30891 and 26650 voxels were identified, respectively. Decreased volume was observed in the interpeduncular nucleus and anterior commissure (data not shown). Thus, VPA-treatment during embryonic development strongly disrupted brain structure and composition, reducing the glutamatergic signal in many brain regions, and differentially affecting GABA neurons in other areas.
Figure 6. Neuroanatomical changes following valproic acid treatment.
A-C. Voxel-intensity analysis of glutamatergic expression. (A-A’) Whole brain maximum intensity projections showing voxels that were significant in cluster analysis (fire) for decreased vglut2a:GFP signal. Dorsal view shown in (A) and sagittal view in (A’). Anterior is to the left. (B) Transverse view of bilateral voxel clusters at anterior tip of statoacoustic ganglia (fire) overlapping vglut2a:GFP transgene expression (green). (C-C’) Affine registered confocal images from individual brains showing vglut2a:GFP expression in untreated (C) and VPA-treated (C’) larvae. Anterior is to the left. In all images, grey staining shows pan-neuronal expression of HuC:Cer transgene. Color-scale (also applies to D,F) indicates relative p-values in voxel-wise comparisons.
D-E. Voxel-intensity analysis of GABAergic expression. (D-D’) Whole brain maximum intensity projections showing voxels that were significant in cluster analysis (fire) for decreased gad1b:RFP signal. Dorsal view shown in (D) and sagittal view in (D’).
Anterior is to the left. (E) Transverse view of bilateral voxel clusters in rhombomere 4 (fire) overlapping gad1b:RFP transgene expression (green). In all images, grey staining shows pan-neuronal expression of huC:Cer transgene.
F. Deformation-field analysis. Dorsal (F) and sagittal (F’) views showing whole brain maximum intensity projections of voxels that were significant in region-wise analysis for increased volume resulting from VPA-treatment. Anterior is to the left. Grey staining shows pan-neuronal expression of huC:Cer transgene. Scale bar 100 μm (all panels).
H:hypothalamus; IPN:interpeduncular nucleus tegmentum; SAg:statoacoustic ganglion;Sp: subpallium; Th:thalamus; VT:ventral thalamus; MTe:medial tectum; R1:rhombomere 1; R4:rhombomere 4; R7:rhombomere 7; TeO:optic tectum; Ce:cerebellum; Po:preoptic area; PLLG:posterior lateral line ganglion; oe:olfactory epithelium; MO:medulla oblongata
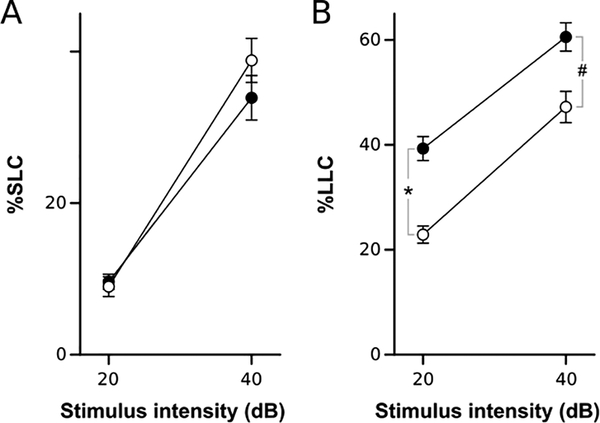
Loss of glutamatergic signal in part of the statoacoustic ganglion lead us to predict that VPA-treated larvae may show reduced escape responses to an acoustic stimulus. Indeed, when probed with auditory stimuli four days after termination of VPA exposure, larvae showed a significant reduction in overall responsiveness (F1,124=11.09, p < 0.001). We found differential effects on escape behavior: long-latency C-start responses were selectively decreased, with no significant effect on short-latency C-start responses (Fig. 7). Thus, anatomical changes were identified with sufficient precision to assess likely effects on behavior.
Figure 7. Valproic acid treatment effect on acoustic startle responsiveness.
A-B Responsiveness to acoustic stimuli. Percents show mean proportion of trials on which larvae executed a short-latency (%SLC, A) or long-latency (%LLC, B) C-start response for two acoustic stimulus intensities. N = 63 larvae per group. # p < 0.001; * p < 0.0001
4. Discussion
Zebrafish is a rapidly emerging model for the study of neurodevelopmental disorders. Although genetic and imaging methods have long been available in this system, progress in elucidating complex, pleiotropic neurodevelopmental phenotypes has been limited by a scarcity of methods to perform detailed brain-wide analysis of neuroanatomical changes. Voxel-based morphometry is a powerful method for investigating anatomical brain abnormalities associated with human neurodevelopmental and psychiatric disorders. Although widely used in human neuroimaging studies, the application of VBM to vertebrate model organisms such as non-human primates and rodents has been limited by the accuracy of brain registration methods [57,58]. Advances in non-linear registration techniques and the construction of digital atlases have now allowed us to develop quantitative methods for voxel, deformation and volume-based analysis of zebrafish brains. This study validates these methods and demonstrates their application to the valproic acid model of ASD. Software for morphometric analysis is freely available from our website (https://science.nichd.nih.gov/confluence/display/burgess/Software). Further, we describe a new neuroanatomical segmentation of the larval brain, based on K-means clustering of transgene expression data. The atlas is available within the download.
Registration techniques have been recently used in zebrafish to map changes in whole-brain activity patterns to a reference atlas, thereby connecting activity with neuroanatomical information [8,59,60]. A similar approach has been applied to analyze brain-wide changes in gene expression in mutant embryos by using optical projection tomography to image chromogenic whole-mount in situ hybridization (WISH) stained samples for voxel-intensity analysis [7]. This method is excellent for analysis of WISH staining, as it utilizes a standard chromogenic staining protocol that produces sensitive and robust signals. However, optical projection tomography requires a specialized imaging platform and WISH necessitates tissue fixation which introduces deformation artifacts [22]. In contrast, the method described in this study uses standard confocal microscopy for live-imaging of transgene expression in wildtype or experimental samples. In principal, the same computational tools can be applied to fixed tissue samples that may be necessary for WISH or immunohistochemistry. Additionally, our method enables detection of shape and volume changes by analyzing deformation fields and measuring the size of brain subdivisions. The major limitation of our method is the time for deriving significance thresholds via permutation testing, typically around 90 minutes using 100 nodes on a computing cluster. However, our software (CobraZ, for Comparative Brain Analysis for Zebrafish) can also run using networked desktop computers, making it feasible to obtain results overnight using PCs available in a typical laboratory setting.
Our analyses use a new neuroanatomical map of 180 subdivisions of the larval brain. Historically, most neuroanatomy atlases have been constructed manually, by identifying regions with salient differences in cell composition or activity in functional studies. However, a particular challenge in the larval zebrafish model is that many brain regions lack conspicuous nuclear organization, making accurate segmentation of distinct regions difficult and subjective. Indeed, the widely used Mueller and Wullimann atlas of the larval brain [61] describes 102 regions, only half of the 195 regions annotated in the adult zebrafish brain [62], reflecting the absence of nuclear boundaries in many areas. To address this problem, we implemented an automated computational procedure that segmented distinct neuroanatomical regions by clustering voxels with similar genetic identity. Our algorithm was derived from K-means clustering, a method that requires a priori knowledge of the number of clusters. Based on the literature, we tested using 100300 clusters, and ultimately selected a map that comprised 180 regions. However this was simply the optimal map using our evaluation criteria and should not be taken as the true number of regions present in the brain. Many of our regions can likely be further subdivided - indeed, alignment of our neuroanatomical atlas with the map of cellular/neuropil zones shows that many regions could be separated at least into cell-rich and neuropil-rich areas. Moreover, when we applied a Bayesian Information Criterion (BIC) to evaluate the optimal number of clusters, we found improvement up to K=800 clusters. We did not pursue this further because of the difficulty in assessing the biological identity and basis for most of these regions. Nevertheless, the procedure accurately delineated known areas and also revealed brain regions that were previously not recognized. Thus by using this unbiased segmentation technique, we obtained a new neuroanatomical map of the zebrafish brain that enabled us to establish separate significance thresholds for different brain regions via permutation analysis, and measure the volume of specific nuclei. We anticipate that this new map will also facilitate mapping of neuronal circuits and comparative studies with mammals.
The three methods that we present for morphometric analysis have complementary strengths. Voxel-intensity comparisons (voxel and cluster analysis) are optimal for identifying changes in the cellular composition of brain scans. Here, we imaged brains with fluorescent reporters in glutamatergic and GABAergic neurons, reporters that are widely expressed. However, the same method can be applied to any population of neurons marked by a transgenic line. Deformation-field and volume-based methods both identify local changes in brain microstructure. An advantage of deformation-field analysis is that it does not rely on the accuracy of the neuroanatomical map and may locate shape changes within brain regions. However, small global changes in scaling may not be detected using this method. Volume-analysis provides a reliable estimate of the actual volume of the entire brain and its subdivisions and has the further advantage that the magnitude of the changes are easily described. We envisage using morphometry to locate changes in brain development for subsequent analysis using molecular genetic methods. As such studies are time-consuming and expensive, a major objective was to provide rigorous control over type I errors. False positives can be avoided by focussing on areas with significant changes that persist after stringent Holm-Bonferroni correction, and that show similar differences across brain hemispheres.
To demonstrate the effectiveness of our methods for detecting neurodevelopmental abnormalities, we analyzed the VPA model of ASD. VPA exposure was first associated with autism based on a high frequency of autism symptoms in patients with fetal valproate syndrome (FVS), a congenital disorder resulting from in utero VPA exposure [63,64]. FVS causes variable symptoms that frequently include facial dysmorphism, heart defects, spina bifida, and developmental delay. In a previous zebrafish study, the effect of treatment with 60 μM VPA was assessed by histopathology and resulted in disruption of normal brain structure characterized by small regions of reduced cellularity [56]. We used a slightly lower dose, and found widespread loss of glutamatergic neuron signal and both gain and loss of GABAergic neuron signal in specific regions. Our imaging conditions could not distinguish whether this reflected a change in the number of neurons of each type, or a change in the level of expression of the fluorescent reporter. However, these changes are consistent with the function of VPA as a histone deacetylase inhibitor that affects the proliferative state of neural progenitor cells [65,66]. Expression of the glutamatergic neuron marker was reduced at the anterior-most region of the statoacoustic ganglion (as defined by a slight gap in neurod:GFP and isl2b:GFP transgene expression prior to the caudal-most part of the anterior lateral line ganglion). This finding led us to successfully predict that behavioral responses to an acoustic stimulus would be diminished. The finding that brain morphometry is sufficiently accurate and sensitive to detect changes that predict alterations in behavior is exciting, offering a new approach to characterizing effects of gene mutations. Additionally, we anticipate that these methods could be used to detect inherent morphological differences in diverse populations or strains of zebrafish, as has been demonstrated in surface-dwelling and cave-adapted Astynax mexicanus, which display changes in the neurodevelopmental program of the forebrain depending on the morph [67].
At a resolution sufficient to resolve individual neurons, morphometry in larval zebrafish uses 5.3 M voxels, comparable to a human T1 MRI study (8 M voxels). Genetic studies in humans are typically limited by the difficulty in recruiting a sufficiently large patient cohort for statistical power. In contrast, large numbers of larvae carrying the same genetic mutation can be readily obtained from zebrafish crosses, such that the main limitation is in data acquisition. At present a whole-brain three channel confocal scan requires around 20 minutes, but with increasing access to selective planar illumination microscopy and automated microscopy platforms, it will become easier to collect very large sample sizes [68,69]. The ability to simultaneously acquire information about whole-brain structure and cellular composition is a key advantage of the zebrafish system. We therefore believe that our new tools for brain morphometry will broadly facilitate the investigation of neurodevelopmental phenotypes by using zebrafish to model the effects of gene mutations and environmental exposures.
Supplementary Material
Supplementary File 1. Zipped folder containing computationally derived neuroanatomical atlas, map of cell-rich brain regions, and a text file with atlas annotations.
Supplemental Figure 1. atoh7 morpholino efficacy
A. Epiflorescent image of retinal ganglion cell arborization (green) in the optic tectum (TeO) of Tg(atoh7:GFP)rw021 larvae injected with atoh7 or control (ctrl) morpholinos (MO).
B. Pigmentation of larvae injected with atoh7 or control morpholinos.
C. O-bend probability of larvae injected with atoh7 or control morpholinos. N=23 & 16 respectively.
Acknowledgements
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute for Child Health and Human Development (NICHD) and utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD.
Abbreviations
- Dpf
days post fertilization
- Hpf
hours post fertilization
- VBM
voxel-based morphometry
- ASD
Autism spectrum disorder
- VPA
valproic acid
Footnotes
Declarations of interest: none
References
- [1].Birnbaum R, Weinberger DR, Genetic insights into the neurodevelopmental origins of schizophrenia., Nat. Rev. Neurosci 18 (2017) 727–740. doi:10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- [2].Geschwind DH, State MW, Gene hunting in autism spectrum disorder: on the path to precision medicine, Lancet Neurol. 14 (2015) 1109–1120. doi:10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gokoolparsadh A, Sutton GJ, Charamko A, Green NFO, Pardy CJ, Voineagu I, Searching for convergent pathways in autism spectrum disorders: insights from human brain transcriptome studies., Cell. Mol. Life Sci. CMLS 73 (2016) 4517–4530. doi:10.1007/s00018-016-2304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kotlar AV, Mercer KB, Zwick ME, Mulle JG, New discoveries in schizophrenia genetics reveal neurobiological pathways: A review of recent findings, Eur. J. Med. Genet 58 (2015) 704–714. doi:10.1016/j.ejmg.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sztainberg Y, Zoghbi HY, Lessons learned from studying syndromic autism spectrum disorders., Nat. Neurosci 19 (2016) 1408–1417. doi:10.1038/nn.4420. [DOI] [PubMed] [Google Scholar]
- [6].Chomiak T, Turner N, Hu B, What We Have Learned about Autism Spectrum Disorder from Valproic Acid., Pathol. Res. Int 2013 (2013) 712758. doi:10.1155/2013/712758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Allalou A, Wu Y, Ghannad-Rezaie M, Eimon PM, Yanik MF, Automated deep-phenotyping of the vertebrate brain., ELife. 6 (2017). doi:10.7554/eLife.23379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Randlett O, Wee CL, Naumann EA, Nnaemeka O, Schoppik D, Fitzgerald JE, Portugues R, Lacoste AM, Riegler C, Engert F, Whole-brain activity mapping onto a zebrafish brain atlas, Nat. Methods 12 (2015) 1039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL, Longitudinal organization of the anterior neural plate and neural tube., Dev. Camb. Engl 121 (1995) 3923–3933. [DOI] [PubMed] [Google Scholar]
- [10].Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH, Zebrafish hox clusters and vertebrate genome evolution., Science. 282 (1998) 1711–1714. [DOI] [PubMed] [Google Scholar]
- [11].Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK, Efficient genome editing in zebrafish using a CRISPR-Cas system., Nat. Biotechnol 31 (2013) 227–229. doi:10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong J-W, Xi JJ, Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos., Cell Res. 23 (2013) 465–472. doi:10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rennekamp AJ, Peterson RT, 15 years of zebrafish chemical screening., Curr. Opin. Chem. Biol. 24 (2015) 58–70. doi:10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wiley DS, Redfield SE, Zon LI, Chemical screening in zebrafish for novel biological and therapeutic discovery., Methods Cell Biol. 138 (2017) 651–679. doi:10.1016/bs.mcb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Furushima K, Murata T, Kiyonari H, Aizawa S, Characterization of Opr deficiency in mouse brain: Subtle defects in dorsomedial telencephalon and medioventral forebrain, Dev. Dyn 232 (2005) 1056–1061. doi:10.1002/dvdy.20253. [DOI] [PubMed] [Google Scholar]
- [16].Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M, Smaller Dendritic Spines, Weaker Synaptic Transmission, but Enhanced Spatial Learning in Mice Lacking Shank1, J. Neurosci 28 (2008) 1697–1708. doi:10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].O’Tuathaigh CMP, Harte M, O’Leary C, O’Sullivan GJ, Blau C, Lai D, Harvey RP, Tighe O, Fagan AJ, Kerskens C, Reynolds GP, Waddington JL, Schizophrenia-related endophenotypes in heterozygous neuregulin-1 ‘knockout’ mice, Eur. J. Neurosci 31 (2010) 349–358. doi:10.1111/j.1460-9568.2009.07069.x. [DOI] [PubMed] [Google Scholar]
- [18].Song C-H, Bernhard D, Bolarinwa C, Hess EJ, Smith Y, Jinnah HA, Subtle microstructural changes of the striatum in a DYT1 knock-in mouse model of dystonia, Neurobiol. Dis 54 (2013) 362–371. doi:10.1016/j.nbd.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hayasaka S, Nichols TE, Combining voxel intensity and cluster extent with permutation test framework, NeuroImage. 23 (2004) 54–63. doi:10.1016/j.neuroimage.2004.04.035. [DOI] [PubMed] [Google Scholar]
- [20].Whitwell JL, Voxel-Based Morphometry: An Automated Technique for Assessing Structural Changes in the Brain, J. Neurosci 29 (2009) 9661–9664. doi:10.1523/JNEUROSCI.2160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Marquart GD, Tabor KM, Brown M, Strykowski JL, Varshney GK, LaFave MC, Mueller T, Burgess SM, Higashijima S-I, Burgess HA, A 3D Searchable Database of Transgenic Zebrafish Gal4 and Cre Lines for Functional Neuroanatomy Studies, Front. Neural Circuits 9 (2015) 78. doi:10.3389/fncir.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marquart GD, Tabor KM, Horstick EJ, Brown M, Geoca AK, Polys NF, Nogare DD, Burgess HA, High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization., GigaScience. 6 (2017) 1–15. doi:10.1093/gigascience/gix056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ronneberger O, Liu K, Rath M, Ruebeta D, Mueller T, Skibbe H, Drayer B, Schmidt T, Filippi A, Nitschke R, Brox T, Burkhardt H, Driever W, ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains, Nat Methods. 9 (2012) 735–42. doi:10.1038/nmeth.2076. [DOI] [PubMed] [Google Scholar]
- [24].Satou C, Kimura Y, Hirata H, Suster ML, Kawakami K, Higashijima S, Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons, Development. 140 (2013) 3927–31. doi:10.1242/dev.099531. [DOI] [PubMed] [Google Scholar]
- [25].Forster D, Arnold-Ammer I, Laurell E, Barker AJ, Fernandes AM, Finger-Baier K, Filosa A, Helmbrecht TO, Kolsch Y, Kuhn E, Robles E, Slanchev K, Thiele TR, Baier H, Kubo F, Genetic targeting and anatomical registration of neuronal populations in the zebrafish brain with a new set of BAC transgenic tools., Sci. Rep 7 (2017) 5230. doi:10.1038/s41598-017-04657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kimura Y, Satou C, Higashijima S, V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord, Development. 135 (2008) 3001–5. [DOI] [PubMed] [Google Scholar]
- [27].Bergeron SA, Carrier N, Li GH, Ahn S, Burgess HA, Gsx1 Expression Defines Neurons Required for Prepulse Inhibition, Mol. Psychiatry (2014). doi:10.1038/mp.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Horstick EJ, Jordan DC, Bergeron SA, Tabor KM, Serpe M, Feldman B, Burgess HA, Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish, 43 (2014) e48. doi:10.1093/nar/gkv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Masai I, Lele Z, Yamaguchi M, Komori A, Nakata A, Nishiwaki Y, Wada H, Tanaka H, Nojima Y, Hammerschmidt M, Wilson SW, Okamoto H, N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites, Development. 130 (2003) 2479–2494. doi:10.1242/dev.00465. [DOI] [PubMed] [Google Scholar]
- [30].Chu J, Haynes RD, Corbel SY, Li P, Gonzalez-Gonzalez E, Burg JS, Ataie NJ, Lam AJ, Cranfill PJ, Baird MA, Davidson MW, Ng H-L, Garcia KC, Contag CH, Shen K, Blau HM, Lin MZ, Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein., Nat. Methods 11 (2014) 572–578. doi:10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hieber V, Dai X, Foreman M, Goldman D, Induction of alpha1-tubulin gene expression during development and regeneration of the fish central nervous system., J. Neurobiol 37 (1998) 429–440. [DOI] [PubMed] [Google Scholar]
- [32].Pittman AJ, Law MY, Chien CB, Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points, Development. 135 (2008) 2865–71. doi:10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Burgess HA, Granato M, Sensorimotor gating in larval zebrafish, J. Neurosci 27 (2007) 4984–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Preibisch S, Saalfeld S, Tomancak P, Globally optimal stitching of tiled 3D microscopic image acquisitions, Bioinformatics. 25 (2009) 1463–5. doi:10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat Methods. 9 (2012) 676–82. doi:10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fowlkes EB, Mallows CL, A Method for Comparing Two Hierarchical Clusterings, J. Am. Stat. Assoc 78 (1983) 553–569. doi:10.1080/01621459.1983.10478008. [Google Scholar]
- [37].Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC, A reproducible evaluation of ANTs similarity metric performance in brain image registration., NeuroImage. 54 (2011) 2033–2044. doi:10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods 9 (2012) 671–675. doi:10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Eklund A, Nichols TE, Knutsson H, Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates, Proc. Natl. Acad. Sci. U. S. A 113 (2016) 7900–7905. doi:10.1073/pnas.1602413113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ng L, Bernard A, Lau C, Overly CC, Dong H-W, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M, An anatomic gene expression atlas of the adult mouse brain., Nat. Neurosci 12 (2009) 356–362. doi:10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- [41].Bohland JW, Bokil H, Pathak SD, Lee C-K, Ng L, Lau C, Kuan C, Hawrylycz M, Mitra PP, Clustering of spatial gene expression patterns in the mouse brain and comparison with classical neuroanatomy., Methods San Diego Calif. 50 (2010) 105–112. doi:10.1016/j.ymeth.2009.09.001. [DOI] [PubMed] [Google Scholar]
- [42].Ji S, Computational genetic neuroanatomy of the developing mouse brain: dimensionality reduction, visualization, and clustering., BMC Bioinformatics. 14 (2013) 222. doi:10.1186/1471-2105-14-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima S, Fetcho J, A structural and functional ground plan for neurons in the hindbrain of zebrafish, Proc Natl Acad Sci U A. 108 (2011) 1164–9. doi:10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O’Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J, Multiplex Targeted Sequencing Identifies Recurrently Mutated Genes in Autism Spectrum Disorders, Science. 338 (2012) 1619–1622. doi:10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH, Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats, Nature. 364 (1993) 717–21. [DOI] [PubMed] [Google Scholar]
- [46].Sztriha L, Vos YJ, Verlind E, Johansen J, Berg B, X-linked hydrocephalus: a novel missense mutation in the L1CAM gene., Pediatr. Neurol 27 (2002) 293–296. [DOI] [PubMed] [Google Scholar]
- [47].Gaser C, Volz H-P, Kiebel S, Riehemann S, Sauer H, Detecting Structural Changes in Whole Brain Based on Nonlinear Deformations—Application to Schizophrenia Research, NeuroImage. 10 (1999) 107–113. doi:10.1006/nimg.1999.0458. [DOI] [PubMed] [Google Scholar]
- [48].Amo R, Aizawa H, Takahoko M, Kobayashi M, Takahashi R, Aoki T, Okamoto H, Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula, J Neurosci. 30 (2010) 1566–74. doi:10.1523/JNEUROSCI.3690-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kay J, Finger-Baier K, Roeser T, Staub W, Baier H, Retinal Ganglion Cell Genesis Requires lakritz, a Zebrafish atonal Homolog, Neuron. 30 (2001) 725–736. [DOI] [PubMed] [Google Scholar]
- [50].Roullet FI, Lai JKY, Foster JA, In utero exposure to valproic acid and autism--a current review of clinical and animal studies., Neurotoxicol. Teratol 36 (2013) 47–56. doi:10.1016/j.ntt.2013.01.004. [DOI] [PubMed] [Google Scholar]
- [51].Nicolini C, Fahnestock M, The valproic acid-induced rodent model of autism., Exp. Neurol 299 (2018) 217–227. doi:10.1016/j.expneurol.2017.04.017. [DOI] [PubMed] [Google Scholar]
- [52].Gogolla N, LeBlanc JJ, Quast KB, Südhof TC, Fagiolini M, Hensch TK, Common circuit defect of excitatory-inhibitory balance in mouse models of autism, J. Neurodev. Disord 1 (2009) 172. doi:10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hussman JP, Suppressed GABAergic inhibition as a common factor in suspected etiologies of autism, J. Autism Dev. Disord 31 (2001) 247–248. [DOI] [PubMed] [Google Scholar]
- [54].Rubenstein JLR, Merzenich MM, Model of autism: increased ratio of excitation/inhibition in key neural systems, Genes Brain Behav. 2 (2003) 255–267. doi:10.1034/j.1601-183X.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K, Neocortical excitation/inhibition balance in information processing and social dysfunction, Nature. 477 (2011) 171–178. doi:10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Beker van Woudenberg A, Snel C, Rijkmans E, de Groot D, Bouma M, Hermsen S, Piersma A, Menke A, Wolterbeek A, Zebrafish embryotoxicity test for developmental (neuro)toxicity: Demo case of an integrated screening approach system using anti-epileptic drugs., Reprod. Toxicol. Elmsford N 49 (2014) 101–116. doi:10.1016/j.reprotox.2014.07.082. [DOI] [PubMed] [Google Scholar]
- [57].Maldjian JA, Shively CA, Nader MA, Friedman DP, Whitlow CT, Multi-Atlas Library for Eliminating Normalization Failures in Non-Human Primates., Neuroinformatics. 14 (2016) 183–190. doi:10.1007/s12021-015-9291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pagani M, Damiano M, Galbusera A, Tsaftaris SA, Gozzi A, Semi-automated registration-based anatomical labelling, voxel based morphometry and cortical thickness mapping of the mouse brain., J. Neurosci. Methods 267 (2016) 62–73. doi:10.1016/j.jneumeth.2016.04.007. [DOI] [PubMed] [Google Scholar]
- [59].Dunn TW, Mu Y, Narayan S, Randlett O, Naumann EA, Yang C-T, Schier AF, Freeman J, Engert F, Ahrens MB, Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion., ELife. 5 (2016) e12741. doi:10.7554/eLife.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Naumann EA, Fitzgerald JE, Dunn TW, Rihel J, Sompolinsky H, Engert F, From Whole-Brain Data to Functional Circuit Models: The Zebrafish Optomotor Response., Cell. 167 (2016) 947–960.e20. doi:10.1016/j.cell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mueller T, Wullimann MF, Atlas of Early Zebrafish Brain Development. A Tool for Molecular Neurogenetics., 1st ed., Elsevier B.V., Amsterdam, 2005. [Google Scholar]
- [62].Wullimann MF, Rupp B, Reichert H, Neuroanatomy of the zebrafish brain: a topological atlas, Birkhäuser, Basel, 1996. [Google Scholar]
- [63].Christensen J, Gronborg TK, Sorensen MJ, Schendel D, Parner ET, Pedersen LH, Vestergaard M, Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism., JAMA. 309 (2013) 1696–1703. doi:10.1001/jama.2013.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Christianson AL, Chesler N, Kromberg JG, Fetal valproate syndrome: clinical and neuro-developmental features in two sibling pairs., Dev. Med. Child Neurol 36 (1994) 361–369. [DOI] [PubMed] [Google Scholar]
- [65].Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T, Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells., EMBO J. 20 (2001) 6969–6978. doi:10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fujimura K, Mitsuhashi T, Shibata S, Shimozato S, Takahashi T, In Utero Exposure to Valproic Acid Induces Neocortical Dysgenesis via Dysregulation of Neural Progenitor Cell Proliferation/Differentiation., J. Neurosci. Off. J. Soc. Neurosci 36 (2016) 10908–10919. doi:10.1523/JNEUROSCI.0229-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Alie A, Devos L, Torres-Paz J, Prunier L, Boulet F, Blin M, Elipot Y, Retaux S, Developmental evolution of the forebrain in cavefish, from natural variations in neuropeptides to behavior., ELife. 7 (2018). doi:10.7554/eLife.32808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chang T-Y, Pardo-Martin C, Allalou A, Wahlby C, Yanik MF, Fully automated cellular-resolution vertebrate screening platform with parallel animal processing., Lab. Chip 12 (2012) 711–716. doi:10.1039/c1lc20849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pitrone PG, Schindelin J, Stuyvenberg L, Preibisch S, Weber M, Eliceiri KW, Huisken J, Tomancak P, OpenSPIM: an open-access light-sheet microscopy platform, Nat Meth. 10 (2013) 598–599. doi:10.1038/nmeth.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1. Zipped folder containing computationally derived neuroanatomical atlas, map of cell-rich brain regions, and a text file with atlas annotations.
Supplemental Figure 1. atoh7 morpholino efficacy
A. Epiflorescent image of retinal ganglion cell arborization (green) in the optic tectum (TeO) of Tg(atoh7:GFP)rw021 larvae injected with atoh7 or control (ctrl) morpholinos (MO).
B. Pigmentation of larvae injected with atoh7 or control morpholinos.
C. O-bend probability of larvae injected with atoh7 or control morpholinos. N=23 & 16 respectively.