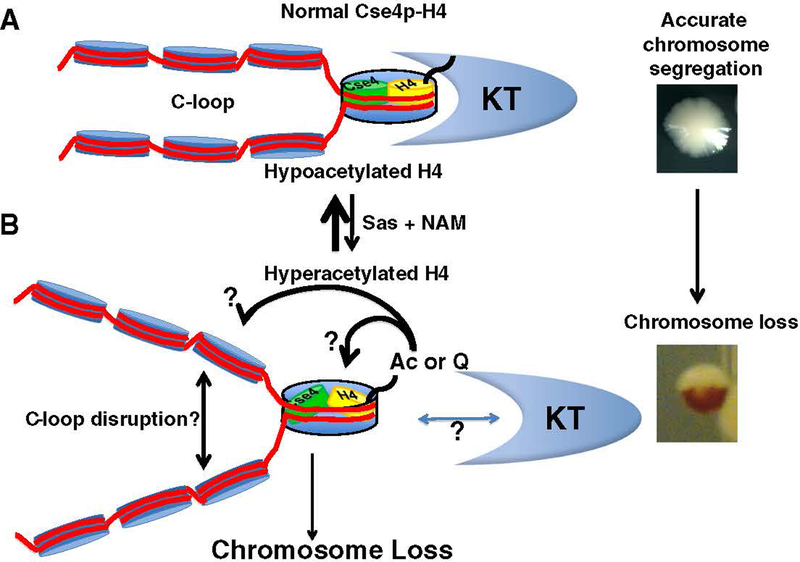
Fig. 1.
Acetylation pattern of centromeric histone H4 affects chromosome segregation. (A) Centromeric histone H4 N-terminal tails are hypoacetylated, which is associated with proper C-loop (light blue nucleosomes to left of Cse4/H4 nucleosome) and kinetochore (KT) assembly to mediate accurate chromosome segregation. Note that the Cse4 nucleosome illustration does not indicate the actual composition of the centromeric nucleosome (refer to the introduction for various models of centromeric nucleosomes). (B) Phenotypes resulting from hyperacetylated centromeric histone H4 at K16. Increased dosage of histone H4K16 acetylation (Ac) by overexpression of Sas2 histone acetyltransferase activity (Sas), growth on nicotinamide (NAM), an inhibitor of nicotinamide adenine dinucleotide (NAD)-dependent deacetylases that act on H4K16, or use of an H4K16Q acetyl-mimic mutant (Q – glutamine) results in chromosome loss that may be caused by disruption of the C-loop and/or improper kinetochore assembly at the centromere (?). (Far right). We use a visual assay to monitor chromosome loss in a strain engineered with a reporter chromosome. Loss of the reporter chromosome results in red sectors in an otherwise white colony. Chromosome loss within the first cell division results in half-sectored red colonies. Refer to Section 2 for details.