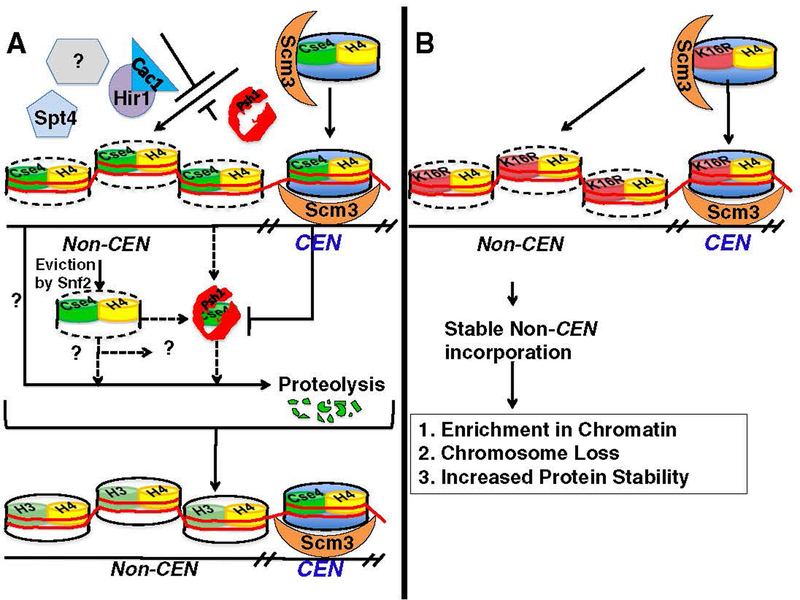
Fig. 2.
Mechanisms that prevent non-centromeric localization of Cse4 to maintain genome stability. (A) Cse4 can localize to non-centromeric regions; however, mis-localization is suppressed by Psh1, Snf2, Cac1-Hir1, Spt4, and other proteins (?). Psh1 is an E3 ubiquitin ligase that mediates proteolytic degradation of Cse4 and excludes its incorporation into non-centromeric DNA. Centromeric incorporation of Cse4 and its interaction with Scm3 is thought to protect Cse4 from proteolysis by Psh1. Snf2 is proposed to remove Cse4 from non-centromeric regions. Cse4 eviction by Snf2 may or may not be mutually exclusive from known (Psh1) and unknown (?) proteolytic pathways. The centromeric nucleosome is depicted in blue, and the non-centromeric regions of Cse4 are denoted by the cylinders with a dotted outline. Note that the Cse4 nucleosome illustration does not indicate the actual composition of the centromeric nucleosome (refer to the introduction for various models of centromeric nucleosomes). (B) Overexpression of cse4K16R leads to its mis-localization to non-centromeric regions. Scm3-mediated centromeric localization of cse4K16R is inferred from complementation of a CSE4 deletion strain by cse4K16R. Stable non-centromeric incorporation of cse4K16R leads to its enrichment in chromatin, chromosome loss, and increased protein stability. Refer to Section 3 for details.