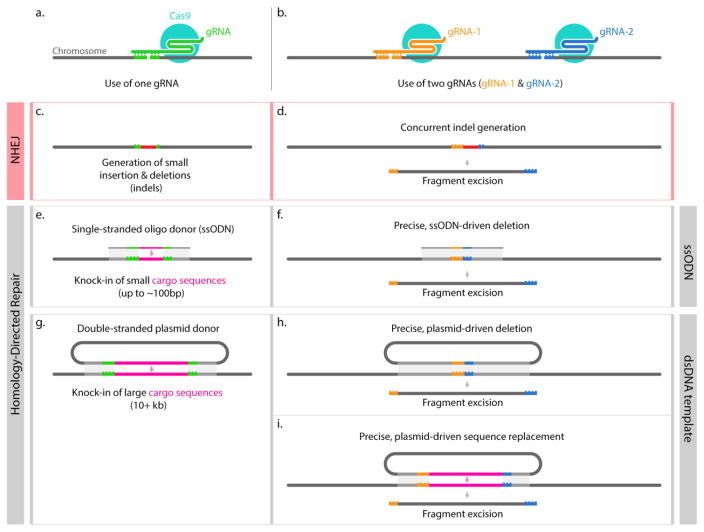
Figure 1. Strategies for genome editing using CRISPR-based tools.
Various gene editing strategies target the genome for dsDNA cleavage with one (panel a, c, e, g), two (panel b, d, f, h–i), or more (not shown) gRNAs. If a localized random mutation is wanted, the use of a single gRNA could lead to small indels (c) through the NHEJ pathway. To obtain large deletions, it is possible to use two gRNAs flanking the DNA fragment to be deleted; a deletion can be achieved without a repair template (d) or by providing a single- or double-stranded DNA that would help bridging the two ends to be fused by HDR (f, h) and obtain predictable sequence contiguity. When the goal is instead the insertion of exogenous cargo sequences, the use of DNA templates helps to do so with base-pair precision at predetermined locations (e, g). When using single-stranded oligodeoxynucleotides (ssODNs) (e) the inserted sequence, limited in size by the oligonucleotide synthesis, can range from few base pairs to ~120 bp (assuming a max oligo length of 200 nt, and homology arms of ~40 nt) (eg.: attP docking sites, FRT or LoxP sequences)[15]. (g) shows instead a strategy where a plasmid is used as a template, in this case, 500–1000 bp homology arms are used, and large DNA sequences can be inserted (10+ Kbp). Lastly, a strategy that combines the approaches in (g) and (h) is diagrammed in (i) where a plasmid is used to simultaneously delete an insert DNA sequences with remarkable accuracy [9,13].