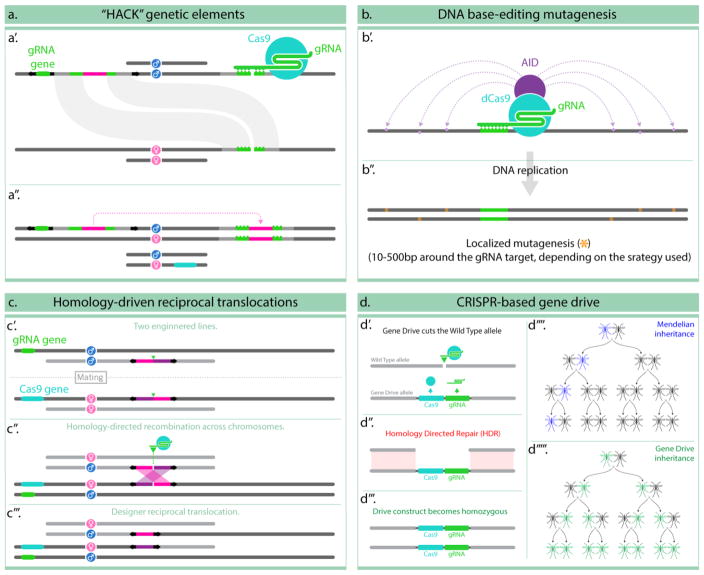
Figure 2. Complex genome editing examples.
In panel (a) and active genetic element with homology arms and a gRNA to a given genomic location is inserted randomly using a transposable element (represented by the black arrows) (a′); further crossing of such transgenic animal with with one carrying a source of Cas9, results in the element copying onto the predetermined location by homology-directed repair (a″) [17]. (b) describes the use of an activation-induced cytidine deaminase (AID), fused to a dead form of Cas9, to target mutagenic DNA base-editing in the neighborhood of the gRNA target site (b′); the endogenous DNA-repair mechanisms repair these mismatches creating occasional mutations in the targeted locus (b″) [19]. (c) describes a strategy that takes advantage of inverted homologous sequences, separated by a gRNA target site, inserted into two separate transgenes (c′); in presence of Cas9 and the appropriate gRNA (in panel (c) they are supplied as transgenes), the gRNA cleaves both locations resulting in four chromosomal fragments with homologous sequences at their ends (c″) which trigger recombination and result in a predictable reciprocal translocation between the darker and the lighter chromosomes carrying the transgenes (c‴) [18]. Lastly, panel (d) describes the use of a transgene containing both a Cas9 and a gRNA genes inserted at the location targeted by the gRNA (d′); this arrangement generates an active genetic element capable of cutting the opposing (wild-type) chromosome (d″) and converting it to the same condition (d‴); when this process happens in the germline of an animal it generated a strong gene drive effect which dramatically modifies the inheritance pattern of the genetic construct from Mendelian (d‴′) to Super-mendelian (d‴″) and can be taken advantage of for population suppression or modification applications.