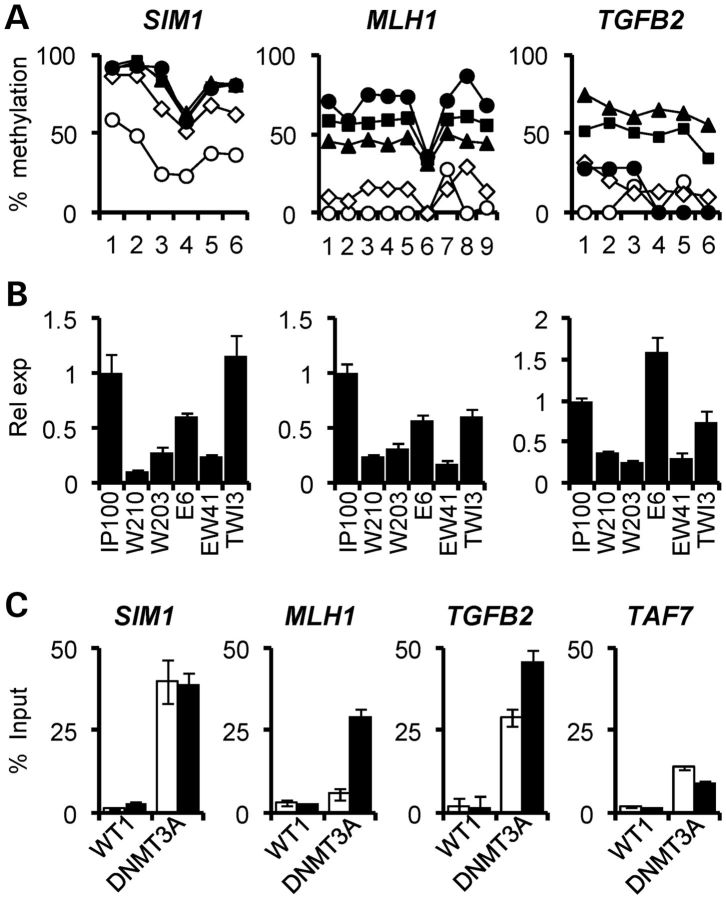
Figure 5.
Increased DNA methylation and gene silencing associated with WT1 expression. (A) High-resolution bisulphite pyrosequencing demonstrating increased DNA methylation at SIM1, MLH1 and TGFB2. Each data point represents an individual CpG dinucleotide. Open circles, IP100; open triangles, E6; black squares, W210; black triangles, W203; black diamonds, EW41. (B) Quantitative gene expression analysis showing silencing of genes hypermethylated in association with high WT1 levels, i.e. W210, W203 and EW41 cells. (C) ChIP analysis showing increased recruitment of DNMT3A at SIM1, MLH1 and TGFB2, which are methylated gene promoters in W210 cells (black bars) compared with IP100 (white bars). The unmethylated TAF7 gene is also shown. Note that WT1 recruitment levels are not altered between IP100 and W210 cells.