Abstract
This study aims to produce transgenic ovine spermatozoa bearing Ossimi sheep growth hormone (Os_GH) cDNA using different methods. The complete coding sequence of Os_GH has been registered in GenBank accession no. KP221575. The sequence of Os_GH cDNA has been subcloned into pmkate2-N expression vectors to construct Os_GH-pmKate2-N vector. Five groups of sperm uptake were submitted. All groups were incubated at 37 °C for 1 h: Control (sperm cells were incubated without vector), Traditional incubation (sperm cells were incubated with vector), Heat shock (sperm cells were incubated with vector at 4 °C for 20 min and heated for 2 min at 42 °C), Heat shock + Dimethyl sulfoxide (DMSO) (sperm cells were incubated with vector and supplemented with 3% of DMSO and then submitted to heat shock regime) and DMSO (sperm cells were incubated with vector and supplemented with 3% DMSO). The sperm genomic DNA in groups was extracted. The Os_GH-pmKate2-N vector was introduced efficiently into the head of sperm cells in all treated groups. Adding DMSO either with or without heat shock increased the sperm uptake. The progressive motility was reduced (P < 0.05) by 29.9% in heat shock group compared to the control. Adding DMSO improved (P < 0.05) the total and progressive motilities by 8.2% and 19.8%, respectively in heat shock group compared to the heat shock group without DMSO. The results documented the ability of ovine spermatozoa to uptake the exogenous vector. Also, sperm incubation with 3% DMSO is the best method to introduce the exogenous vector into spermatozoa without notable adverse effects on sperm motilities.
Keywords: Ovine growth hormone, pmKate2-N expression vector, Sperm uptake
1. Introduction
The sperm mediated gene transfer (SMGT) is a technique that could be used to produce transgenic animals [26]. This technique utilizes the capacity of the spermatozoa to uptake the exogenous DNA and then introduces it into the oocyte during fertilization [11]. Growth hormone (GH) is a single-chain polypeptide hormone synthesized and secreted by the pituitary gland under hypothalamic control [16], [28]. Growth hormone causes an increase in muscles and bone growth and milk production and decrease in fatness [19]. Concentration of blood GH had increased in cows genetically selected for high milk production [2] and in sheep selected for low fat [5]. Transgenic mice, rabbits, sheep and pigs have been used as models to investigate the growth performance of mammals [21], [29]. The blood plasma growth hormone was 10–20-fold in transgenic GH-sheep compared to the control [15]. The objective of the present study was to investigate the capacity of ovine spermatozoa to uptake the exogenous Os_GH-pmKate2-N expression vector using different methods and its impact on sperm motility.
2. Materials and methods
2.1. Semen collection, evaluation and cryopreservation
A total of 18 semen samples were collected from three adult Rahmani rams twice a week during 9 weeks by an artificial vagina at Animal Production Dept., Faculty of Agriculture, Cairo University. The semen advanced motility (%), abnormalities (%), live sperm (%) and sperm concentration (×109/ml) were evaluated according to Hafez and Hafez [7]. After evaluation, the semen samples were diluted with Tris-based extender to give a final sperm concentration of 250 × 106/ml as described previously by Fukui et al., [6]. Tris-based extender contains 297.58 mM Tris, 96.32 mM citric acid, 82.66 mM fructose, 5% (v/v) glycerol, 15% (v/v) egg yolk and 500 µl/ml gentamycin according to Vivanco and Alarcon [27]. The diluted semen was gradually cooled to 4 °C for 2–3 h, then packed in 0.25 ml straws and exposed to the vapor of liquid nitrogen for 3–4 min and plunged into liquid nitrogen [14].
2.2. Isolation of ovine growth hormone cDNA
Total RNA was isolated from pituitary gland of Ossimi sheep (Os) using Booze reagent kit (Bioflux®) and converted into cDNA using Oligo dT18 and RevertAid First Strand cDNA Synthesis Kit (Fermentas, Canada). The Os_GH cDNA was amplified using designed forward primer 5′-GCTCACCAGCTATGAT GGCTG-3′ and reverse primer 5′-TGGCAACTAGAAGGCGCAGCT-3′. The PCR reaction mixture consisted of 1 μl of 10× buffer containing MgCl2 (25 mM), 1 μl of dNTPase mixture (200 μM), 1 μl of F-primer, 1 μl of R-primer, 0.2 μl of Taq polymerase (1 U/μl), 1 μl of Os_GH cDNA and water nuclease-free up to 10 μl. The PCR condition was denaturation for 1 min at 94 °C, annealing for 2 min at 63 °C and extension for 3 min at 72 °C for 29 cycles and final extension at 72 °C for 7 min. The PCR product was visualized using 1% agarose gel stained with ethidium bromide through 1× TAE buffer at 100 V for 1 h with ladder marker range from 100 to 10,000 bp (New England, UK).
2.3. Cloning and expression vectors construction
The visualized band of Os_GH cDNA was extracted from gel using GeneElute™ Gel Extraction Kit (Fermentas, USA) and cloned into pTZ57R/T cloning vector (Fig. 1) according to the kit instructions (2886 bp, Fermentas, USA) as follows: 3 μl of pTZ57R/T vector, 2.5 μl of Os_GH cDNA, 6 μl of 10× ligase buffer, 1 μl T4 DNA ligase and 17.5 μl sterile water in total 30 μl reaction mixture and incubated at 22 °C for 1 h for ligation. The ligated product was transformed into DH10B cells according to the kit instructions (Fermentas, USA). The constructed Os_GH-PTZ57R/T vector was extracted from DH10B cells using GeneJET plasmid Miniprep kit. The efficiency of transformation was determined using PCR and electrophoresis.
Fig. 1.
Map of the pTZ57RT cloning vector. rep (pMB1), A replicon (rep) from the pMBI plasmid is responsible for the replication; bla (ApR), β-lactamase gene conferring resistance to ampicillin; LacZ α-peptide, Blue/white screening of recombinant clones; Phage f1 origin, Synthesis of a single-stranded DNA; Cloning site, 3′-ddT tailed DNA ends for ligation with insert; T7 promoter, In vitro transcription of insert DNA with T7 RNA polymerase [18].
The pmKate2-N is a mammalian expression vector (Fig. 2) that encodes far-red fluorescent protein (4700 bp, Evrogen, Cat. No. FP182, Russia). The Os_GH-pTZ57R/T cloning vector and pmKate2-N expression vector digested with two restriction enzymes SacI and SacII (Jean Bioscience GmbH, Germany). The digested Os_GH cDNA sequence was ligated into the digested pmKate2-N expression vector in reaction volume 20 µl containing 1 µl of digested pmKate2-N expression vector, 2.5 µl of digested Os_GH cDNA, 2 µl of 10× ligase buffer, 1 µl T4 DNA ligase, and 13.5 µl sterile water and was incubated at 22 °C for 1 h for ligation. The constructed Os_GH-pmKate2-N vector was transformed into DH10B cells according to the kit instructions (Fermentas, USA) and extracted using GeneJET plasmid Miniprep kit. The transformation was confirmed by PCR and electrophoresis. The construction method of Os_GH-pmKate2-N expression vector is summarized in Fig. 3.
Fig. 2.
The back bone of pmKate2-N expression vector. MCS, Multiple cloning site for cloned insertion; mKate2, far-red fluorescent protein; PCMV IE, immediate early promoter of cytomegalovirus; SV40 ori, origin for replication in mammalian cells; pUC ori, origin of replication for propagation in E. coli; f1 ori, origin for single-stranded DNA production; SV40 poly A, polyadenylation signals. PSV40, early promoter provides neomycin resistance gene (Neor) expression to select stably transfected eukaryotic cells using G418; P, Bacterial promoter provides kanamycin resistance gene expression (Kanr) in E. coli; Kanr/Neor, gene is linked with herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signals [25].
Fig. 3.
The construction method of Os_GH-pmKate2-N mammalian expression vector. The Os_GH-pTZ57R/T cloning vector and pmKate2-N expression vector were digested with SacI and SacII restriction enzymes to make sticky ends in both Os_GH cDNA fragment and pmKate2-N vector. The digested Os_GH cDNA fragment was ligated into the digested pmKate2-N vector. Finally, the constructed Os_GH-pmKate2-N vector was transformed into DH10B cells for amplification (adapted by authors).
2.4. Semen preparation
The cryopreserved semen straws were thawed by immersion in water bath at 37 °C for 1 min. The pooled semen samples (7 straws) were kept in water bath at 37 °C for 5 min and then assayed using Computer Assisted Sperm Analysis (CASA) instrument (SpermVision™ software MiniTube, version 3.0, USA) connected to Olympus BX 51 microscope (Olympus, Japan). The sperm motion parameters recorded were total motility (%), progressive motility (%), distance curved line (DCL, µm), distance average path (DAP, µm), distance straight line (DSL, µm), velocity average line (VAP, µm/s), velocity curved line (VCL, µm/s), velocity straight line (VSL, µm/s), straightness (STR = VSL/VAP, %), linearity (LIN = VSL/VCL, %), wobble (WOB = VAP/VCL), amplitude of lateral head displacement (ALH, µm) and beat cross frequency (BCF, H2). The pooled semen that has high percentage of advanced motility, live sperm, straightness and linearity was washed three times using 500 µl of 0.9 NaCl by centrifugation at 4000 rpm for 5 min to remove the seminal plasma that contains inhibitory factors. The seminal plasma inhibitory factors may compete with exogenous DNA for the same binding region on the sperm surface according to [12]. The washed semen was re-suspended with Tris extender to a final concentration of 40 × 106 spermatozoa and then assayed using CASA.
2.5. Sperm uptake experiments
A total of 160 μl ovine sperm cell suspension (40 × 106 sperm) was incubated with 200 ng of Os_GH-pmkate2-N expression vector according to Kuznetsov et al. [10]. A total of five groups were submitted in sperm uptake experiments using pmKate2-N expression vector; control group 1 (sperm cells incubated at 37 °C for 1 h without vector), Traditional incubation group 2 (sperm cells incubated at 37 °C for 1 h with vector), Heat shock group 3 (sperm cells incubated with vector at 4 °C for 20 min and heated for 42 °C for 2 min then incubated at 37 °C for 1 h), Heat shock + Dimethyl sulfoxide (DMSO) group 4 (sperm cells incubated with vector and supplemented with 1% of DMSO, then vortexed and incubated at room temperature for 10 min and supplemented with 2% DMSO and then submitted to heat shock regime as group 3) and DMSO group 5 (sperm cells incubated with vector and submitted to DMSO regime as group 4 without heat shock then incubated at 37 °C for 1 h). All incubated groups were washed three times with 500 µl of 0.9% saline by centrifugation at 4000 rpm for 5 min to remove the unbind Os_GH-pmKate2-N vector. The sperm motion parameters were performed for washed semen groups using CASA.
2.6. Recognition of pmKate2-N vector in the spermatozoa
The genomic DNA of the spermatozoa was extracted according to Jerzy et al. [9]. The specific primer for pmKate2-N red fusion protein sequence has been designed to confirm the presence of pmKate2-N vector in the extracted genomic DNA of sperm cells; the forward primer was 5′-CCACTCGCTCGACTAATTCC-3′ and the reverse primer was 5′-GATCCCTCCAGCGTCATAGA-3′. The PCR mixture contained 1 µl of 10 × buffer containing MgCl2 (25 mM), 1 µl of dNTPase (dATP, dTTP, dCTP and dGTP) (200 µM), 1 µl of forward primer, 1 µl of reverse primer, 0.2 µl of Taq polymerase (1 U/µl), 1 µl (50 ng) of total semen DNA and water nuclease-free up to 10 µl. The PCR conditions were denaturation for 1 min at 94 °C, annealing for 30 s at 59 °C, and extension for 3 min at 72 °C, and then final extension at 72 °C for 5 min after 35 cycles. The PCR product of mKate2 red fusion sequence (690 bp) was electrophoresed and visualized as mentioned above.
3. Statistical analysis
Data of sperm motion parameters after sperm uptake experiments were analyzed using the general linear model [20] according to the following model:
Yij = the observation ij.
µ = Overall mean.
Ti = Treatment (i = 1, Control; i = 2, Traditional incubation, i = 3, Heat shock, i = 4, Heat shock + DMSO, i = 5, DMSO only).
Eijk = Experimental error, ij observation assumed to be randomly distributed (0 − б2).
The differences among means were tested [3].
4. Results and discussion
4.1. Growth hormone cDNA isolation and subcloning
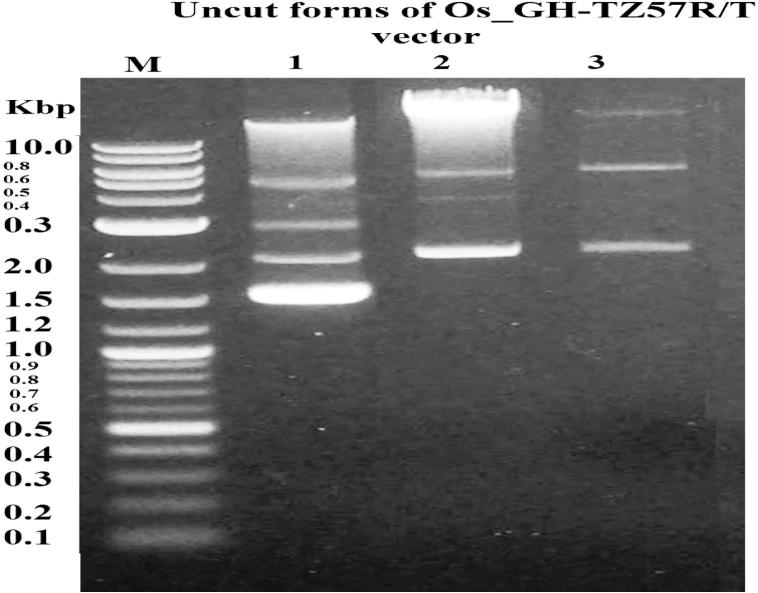
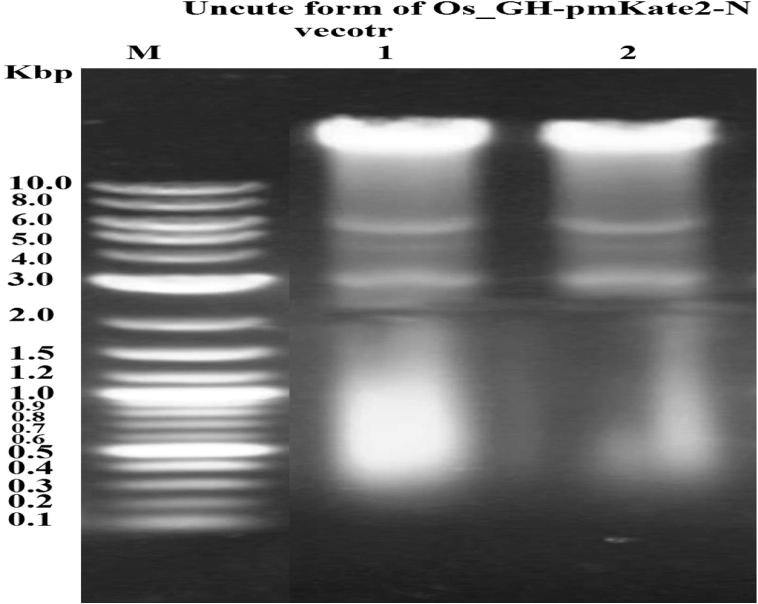
The electrophoretic band of Os_GH cDNA was 690 bp (Fig. 4). The uncut forms of Os_GH-TZ57R/T (3575 bp) and Os_GH-pmKate2-N (5390 bp) vectors are shown in Fig. 5, Fig. 6, respectively. The PCR electrophoresis bands of Os_GHTZ57R/T cloning and Os_GH-pmKate2-N expression vectors were 690 bp (Fig. 7, Fig. 8, respectively) that might indicate that the sequence of Os_GH cDNA was subcloned into pTZ57R/T and pmKate2-N vectors successfully.
Fig. 4.
Electrophoretic pattern of Os_GH cDNA band. M refers to DNA marker given in Kbp; Os_GH cDNA, Ossimi sheep growth hormone cDNA; 1, 2 and 3 are the samples of Os_GH cDNA.
Fig. 5.
Electrophoretic pattern of uncut forms of Os_GH-TZ57RT cloning vector. M refers to DNA marker given in Kbp; 1, 2 and 3 are the samples of uncut Os_GH-TZ57R/T cloning vector.
Fig. 6.
Electrophoretic pattern of uncut forms of Os_GH-pmKate2-N expression vector. M refers to DNA marker given in Kbp; 1 and 2 are the samples of uncut forms of Os_GH-pmKate2-N expression vector.
Fig. 7.
Electrophoretic pattern of Os_GH-TZ57RT cloning vector (PCR). M refers to DNA marker given in Kbp; 1, 2 and 3 are the samples of Os_GH-TZ57R/T cloning vector.
Fig. 8.
Electrophoretic pattern of Os_GH-pmKate2-N expression vector band (PCR, 740 bp). M refers to DNA marker given in Kbp; 1, 2 and 3 are the samples of Os_GH-pmKate2-N vector.
4.2. Sperm uptake
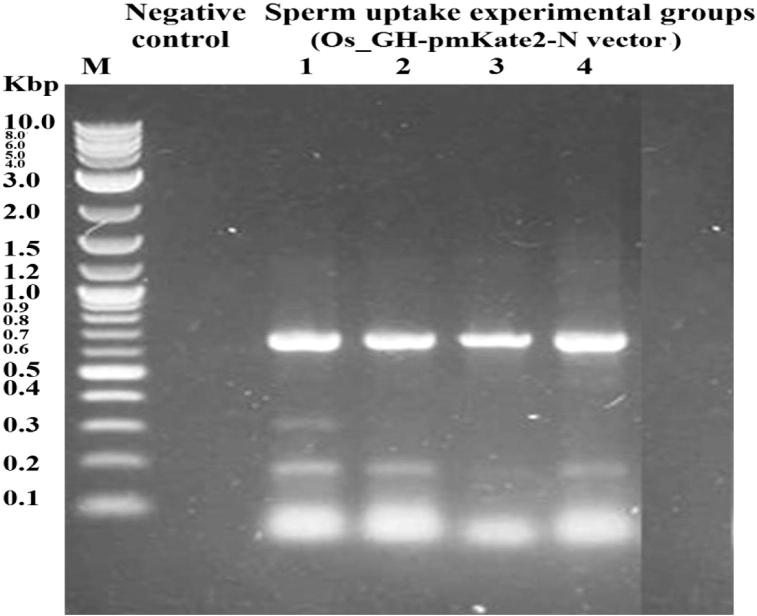
The physical characteristics of Rahmani semen samples before cryopreservation were 84.2% for advanced motility, 369 × 109 for sperm concentration, 4.8% for sperm abnormalities and 81.8% for live sperm. All tested semen characteristics in the present study were in normal range as reported previously [24]. After cryopreservation, the advanced sperm motility was reduced from 84.2% to 70.2%. The PCR electrophoresis of spermatozoa after incubation with Os_GH-pmKate2-N expression vector (Fig. 9) illustrated that Os_GH-pmKate2-N expression vector was introduced efficiently into the head of spermatozoa in all treated groups. The Os_GH-pmKate2-N vector was spontaneously uptaken by spermatozoa incubated at 37 °C for 1 h.
Fig. 9.
Electrophoretic pattern of Os_GH-pmkate2-N vector after sperm uptake using mkate primer (PCR, 690 bp). Line1, l M refers to DNA marker given in Kbp, Line2, Negative control (spermatozoa incubated at 37 °C/1 h without vector), Line3 (1), Traditional incubation group (spermatozoa incubated at 37 °C/1 h with vector), Line4 (2), Heat shock group (spermatozoa incubated at 4 °C/20 min with vector, then at 42 °C/2 min and finally at 37 °C/1 h), line5 (3), Heat shock + DMSO group (spermatozoa supplemented with 3% DMSO and incubated with vector at 4 °C/20 min, then at 42 °C/2 min and finally at 37 °C/1 h) and line6 (4), DMSO group (spermatozoa supplemented with 3% DMSO and incubated with vector at 37 °C/1 h).
The enhancement effect of DMSO on sperm uptake is supported by the results reported by Jacob and Herschler [8], where the DMSO has physiological and technical characteristics of replacing the water in living cells, which interferes with the production of intracellular free oxygen radicals, increasing the permeability of cell membranes, so all molecules are transported across cell membranes of sperm. In the present study, the results of sperm uptake efficiency are supported by Kuznetsov et al. [10], where transgenic rabbits were produced using sperm mediated gene transfer.
4.3. Computer assisted sperm analysis (CASA)
The cryopreserved semen straws have been pooled and washed to remove the seminal plasma inhibitory factors that might prevent the dangerous molecules such as foreign DNA from gaining access to spermatozoa [12]. The seminal plasma inhibitory factors such as Polyamines are basic molecules presented in the most mammalian species and strongly binding to acidic or negatively charged molecules, such as DNA [23].
A second seminal plasma inhibitory factor is glycosaminoglycans that might interfere with the binding of foreign DNA to spermatozoa by competing for the same binding sites [13]. The incubation effect of Os_GH-pmKate2-N vector and treatments on sperm motility traits is shown in Table 1. The total sperm motility was reduced significantly by 5.5% in spermatozoa that incubated with vector at 37 °C for 1 h and by 15.9% in Heat shock group compared to the control (60.5% and 53.8% vs. 64.0%, respectively). Adding DMSO in Heat shock group improved (P < 0.05) the total sperm motility by 8.2% compared to the Heat shock without DMSO (58.2% vs. 53.8%, respectively). In addition, the total sperm motility was increased (P < 0.05) in DMSO group than the other groups. On the other hand, the progressive motility was decreased (P < 0.05) by 13.1% in incubated spermatozoa at 37 °C/1 h compared to the control (45.0% vs. 51.8%, respectively). The Heat shock group had a significant adverse effect on the progressive motility reduced by 29.9% compared to the control (36.3% vs. 51.8%, respectively).
Table 1.
Effect of Os_GH-pmKate2-N vector uptake on sperm motility (mean ± SEM).
| Parameters | Control (37 °C/1 h) | Os_GH-pmKate2-N expression vector groups |
|||
|---|---|---|---|---|---|
| Traditional incubationA | Heat shockB | Heat shockC + DMSO | DMSOD | ||
| Total motility (%) | 64.0 ± 0.8ab | 60.5 ± 0.8c | 53.8 ± 0.8e | 58.2 ± 0.7d | 67.0 ± 0.7a |
| Progressive motility (%) | 51.8 ± 0.8b | 45.0 ± 0.9c | 36.3 ± 0.1e | 43.5 ± 0.6d | 54.9 ± 1.0a |
| DAP (μm) | 23.8 ± 0.3cd | 23.9 ± 0.4cd | 25.0 ± 0.3ab | 25.2 ± 0.2a | 24.7 ± 0.4bc |
| DCL (μm) | 38.0 ± 0.4cd | 38.0 ± 0.5cd | 40.4c ± 0.4ab | 41.8 ± 0.4a | 38.7 ± 0.5c |
| DSL (μm) | 20.6 ± 0.3d | 20.8 ± 0.4cd | 22.0 ± 0.3a | 21.7 ± 0.2ab | 21.5 ± 0.3bc |
| VAP (μm/s) | 51.8 ± 0.6cd | 51.8 ± 0.8cd | 54.2 ± 0.7a | 54.1 ± 0.5ab | 52.9 ± 0.8c |
| VCL (μm/s) | 82.6 ± 0.9cd | 82.4 ± 1.2cd | 87.2 ± 0.8b | 89.8 ± 0.9a | 83.0 ± 1.2c |
| VSL (μm/s) | 44.7 ± 0.6de | 45.1 ± 0.8cd | 47.5 ± 0.6a | 46.6 ± 0.5ab | 46.0 ± 0.8bc |
| STR (VSL/VAP, %) | 0.86 ± 0.0ab | 0.86 ± 0.0ab | 0.87 ± 0.0a | 0.85 ± 0.0bc | 0.86 ± 0.0ab |
| LIN (VSL/CCL, %) | 0.54 ± 0.0bc | 0.54 ± 0.0ab | 0.53 ± 0.0cd | 0.51 ± 0.0e | 0.55 ± 0.0a |
| WOB (VAP/VCL) | 0.62 ± 0.0b | 0.62 ± 0.0b | 0.61c ± 0.0bc | 0.60 ± 0.0d | 0.63 ± 0.0a |
| ALH (μm) | 2.9 ± 0.0a | 2.8 ± 0.0b | 2.9 ± 0.0a | 2.8 ± 0.0b | 2.8 ± 0.0b |
| BCF (H2) | 28.8 ± 0.2b | 29.2 ± 0.3ab | 29.7 ± 0.3a | 28.5 ± 0.2b | 28.6 ± 0.2b |
Means having different superscript letters (a, b, c, d, e) within the same row differ (P < 0.05). DAP, Distance Average Path (microns); DCL, Distance Curved Line (microns); DSL, Distance Straight Line (microns); VAP, Velocity Average Line (microns/sec); VCL, Velocity Curved Line (microns/sec); VSL, Velocity Straight Line (microns/sec); STR, Straightness (VSL/VAP); LIN, Linearity (VSL/VCL); WOB, side to side movement of the sperm head (Wobble, VAP/VCL); ALH, Amplitude of Lateral Head Displacement (microns); BCF, Beat Cross Frequency (H2).
Traditional incubation group, spermatozoa incubated at 37 °C/1 h with vector.
Heat shock group, spermatozoa incubated with vector at 4 °C/20 min then at 42 °C/2 min and finally at 37 °C/1 h.
Heat shock + DMSO group, spermatozoa supplemented with 3% DMSO and incubated with vector at 4 °C/20 min then 42 °C/2 min and finally at 37 °C/1 h.
DMSO group, spermatozoa supplemented with 3% DMSO and incubated with vector at 37 °C/1 h.
In Heat shock group, the progressive motility was enhanced by 19.8% when adding 3% DMSO compared to Heat shock group without adding DMSO (43.5% vs. 36.3%, respectively). Furthermore, the progressive sperm motility was higher (P < 0.05) in DMSO group than other treated groups. This enhancement in total and progressive motilities may be due to the effect of the physiological characteristics of DMSO. The percentage of sperm STR was significantly (P < 0.05) higher in Heat shock group than Heat shock + DMSO but did not differ significantly with other groups. In DMSO group, the WOB value was higher (P < 0.05) than Heat shock + DMSO group and did not differ (P < 0.05) compared to the other treated groups.
The reduction in Wob value in Heat shock group with or without DMSO may be attributed to the adverse effect of heat shock on the movement of head sperm. In traditional incubation and heat shock groups, the value of BCF was increased (P < 0.05) compared to other experimental groups. The values of VSL, VCL, ALH, STR and LIN were increased (P < 0.05) in all treated groups compared to the control. The semen parameters such as VSL, VCL, ALH, STR and LIN were positively correlated with bull fertility [4], [17]. The motility and velocity parameters of spermatozoa reflect their mitochondrial function and energy status indirectly. The higher values of VCL and ALH indicate that there is a major bending of the mid piece and large amplitude of lateral head displacement. This signifies the hyper-activation of the spermatozoa that in turn implies high energy state of the spermatozoa, which is essential for sperm penetration through cervical mucus, zona pellucida, fuse with the oocytes, and successful fertilization [1].
According to the current study, the sperm hyperactivity was observed in Heat shock, Heat shock + DMSO and DMSO groups. Sebastian et al. [22] reported that some sperm motions such as VSL (44.06% vs. 64.80%), VAP (69.39% vs. 89.31%), LIN (37.96% vs. 52.86%), and SRT (61.68 vs. 71.69) were reduced (P < 0.05) after semen incubation with exogenous DNA compared to the control group. On the other hand, they concluded that such reduction in VSL, VAP, LIN, and SRT did not affect the ability of the sperm to fertilize the oocyte in vitro.
5. Conclusions
The Os_GH-pmKate2-N expression vector has successfully uptaken by ovine spermatozoa in all groups: Traditional incubation group, Heat shock group, Heat shock + DMSO group and DMSO group without significant adverse effects on sperm motion.
Authors' contributions
Waleid Shakweer, design of experimental trials, writing the paper, construction of cloning and expression vector, semen collection, evaluation, cryopreservation and sperm uptake experiment and Computer assisted sperm analysis (CASA). Yassein Hafez, design of experimental trials, statistical analysis of results and paper revision. Ashraf EL-Sayed, paper revision. Ibrahim Awadalla and Mamdouh Mohamed, providing the animals for semen collection and animals care.
Acknowledgments
Thanks to Dr. Mamdouh Sharaf El-Deen, Professor of Animal Production, Animal Production Department, Faculty of Agriculture, Cairo University and chairman of Cairo Poultry Company (CPC), for his financial support of this work. Thanks to Dr. Alexander Krivoruchko, Head of Regional Center of Veterinarian Medicine, of Stavropol State, Agrarian University, Russia, for gifting us the pmKat2-N mammalian expression vector.
Footnotes
Peer review under responsibility of National Research Center, Egypt.
References
- 1.Aitken R.J., Sutton M., Warner P., Richardson D.W. J. Reprod. Fertil. 1985;2:441–449. doi: 10.1530/jrf.0.0730441. [DOI] [PubMed] [Google Scholar]
- 2.Bonczek R.R., Young C.W., Wheaton J.E., Miller K.P. J. Dairy Sci. 1988;71:2470–2479. [Google Scholar]
- 3.Duncan D.B. Biometrics. 1955;11:1–15. [Google Scholar]
- 4.Farrell P.B., Foote R.H., McArdle M.M., Trouern V.L., Tardif A.L. J. Androl. 1996;3:293–300. [PubMed] [Google Scholar]
- 5.Francis S.M., Veenvliet B.A., Littlejohn R.P., Stuart S.K., Suttie J.M. Anim. Prod. 1995;55:272–274. [Google Scholar]
- 6.Fukui Y., Kohno H., Togari T., Hiwasa M., Okabe K. J. Reprod. Dev. 2008;4:286–289. doi: 10.1262/jrd.20004. [DOI] [PubMed] [Google Scholar]
- 7.Hafez B., Hafez E.S.E. seventh ed. Wilkens; New York: 2000. Reproduction in Farm Animals. [Google Scholar]
- 8.Jacob S.W., Herschler R. Cryobiology. 1986;1:14–27. doi: 10.1016/0011-2240(86)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Jerzy R., Miroslaw P., Zmudzinski J.F. Bull. Veterin. Inst. Pulawy. 2003;47:71–75. [Google Scholar]
- 10.Kuznetsov A.V., Kuznetsova I.V., Schit I.Y.U. Mol. Reprod. Dev. 2000;56:292–297. doi: 10.1002/(SICI)1098-2795(200006)56:2+<292::AID-MRD18>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 11.Lavitrano M., Camaioni A., Fazio V.M., Dolci S., Farace M.G., Spadafora C. Cell. 1989;57:717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- 12.Lavitrano M., French D., Zani M., Frati L., Spadafora C. Mol. Reprod. Dev. 2006;3:161–169. doi: 10.1002/mrd.1080310302. [DOI] [PubMed] [Google Scholar]
- 13.Lee C.N., Handrow R.R., Lenz R.W., Ax R.L. Gamete Res. 1985;12:345–355. [Google Scholar]
- 14.Matsuoka T., Imai H., Kohno H., Fukui Y. J. Reprod. 2006;52:675–683. doi: 10.1262/jrd.18033. [DOI] [PubMed] [Google Scholar]
- 15.Murray J.D., Nancarrow C.D., Marshall J.T., Hazelton I.G., Ward K.A. Reprod. Fertil. Dev. 1989;1:147–155. doi: 10.1071/rd9890147. [DOI] [PubMed] [Google Scholar]
- 16.Nicoll C.S., Tarpey J.F., Mayer G.L., Russell S. Am. Zool. 1986;4:965–983. [Google Scholar]
- 17.Perumal P., Selvaraju S., Selvakumar S. Reprod. Domest. Anim. 2011;4:636–641. doi: 10.1111/j.1439-0531.2010.01719.x. [DOI] [PubMed] [Google Scholar]
- 18.Rand K.N. Tech. Tips Online. 1996;1:23–24. [Google Scholar]
- 19.Revol A., Garza R.M.D.L., Hernández M.V., Aguilera C., Barrera S.H., Mendoza R. Comp. Biochem. Physiol. 2005;4:423–429. doi: 10.1016/j.cbpb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 20.SAS, SAS User’s Guide for Personal Computers, SAS Institute Inc., Cary, USA, 2000.
- 21.Seamark R.E. In: Manipulating Pig Production. Barnett J.L., Batterham E.S., Cronin G.M., Hansen C., Hemsworth P.H., Hennessy D.P., Hughes P.E., Johnston N.E., King R.H., editors. VIP Printing Pty. Ltd.; Australia: 1987. pp. 165–170. [Google Scholar]
- 22.Sebastian C., Alfonso G., Joaquin G. Mol. Reprod. Dev. 2010;77:687–698. [Google Scholar]
- 23.Setchell B.P., Brooks D.E. In: The Physiology of Reproduction. Knobil E., Neill J.D., editors. Raven Press; New York: 1988. pp. 828–836. [Google Scholar]
- 24.W.M.E. Shakweer, M.Sc. Thesis, Animal Production Department, Faculty of Agriculture, Cairo University, 2008.
- 25.Shcherbo D., Murphy C.S., Ermakova G.V., Solovieva E.A., Chepurnykh T.V., Shcheglov A.S., Verkhusha V.V., Pletnev V.Z., Hazelwood K.L., Roche P.M., Lukyanov S., Zaraisky A.G., Davidson M.W., Chudakov D.M. Biochemistry. 2009;15:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spadafora C. BioEssays. 1998;20:955–964. doi: 10.1002/(SICI)1521-1878(199811)20:11<955::AID-BIES11>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Vivanco H.W., Alarcon V.P. Proc. Western Section Am. Soc. Anim. Sci. 1987;38:237–239. [Google Scholar]
- 28.Wallis M. J. Mol. Evol. 1994;38:619–627. doi: 10.1007/BF00175882. [DOI] [PubMed] [Google Scholar]
- 29.Wieghart M., Hoover J.L., McGrane M.M., Hanson R.W., Rottman F.M., Holtzman S.H., Wagne T.E., Pinkert C.A. J. Reprod. Fertil. 1990;41:89–96. [PubMed] [Google Scholar]