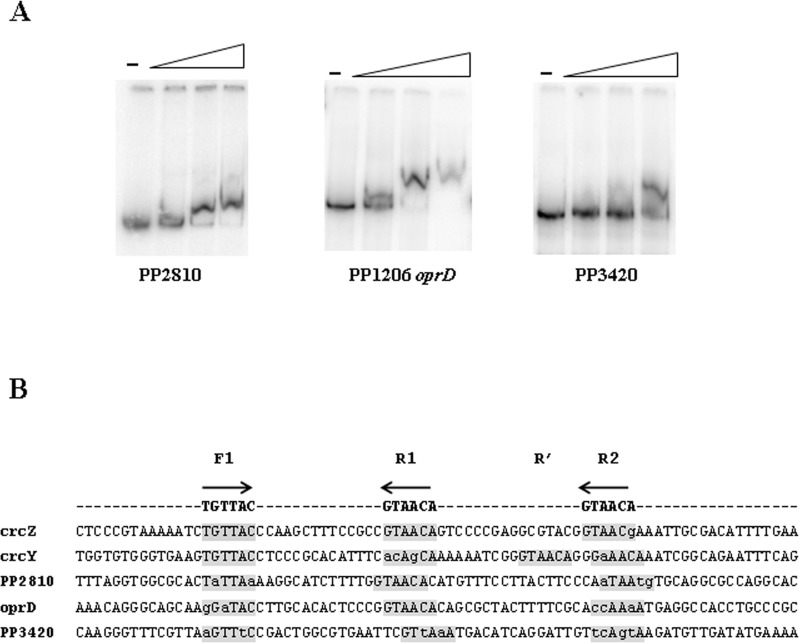
Fig 1.
(A) Electrophoretic mobility shift assay (EMSA) for CbrB binding on PP2810, PP1206 (oprD) and PP3420 promoter regions. Band shift observed in linear dsDNA fragments containing the promoter regions in the presence of increasing amounts of CbrB (0, 1, 1.5 and 2 μM for PP2810 and 0, 0.5, 1 and 2 μM of CbrB for PP1206 and PP3420). A representative assay of three replicas performed is shown. (B) Sequence alignment of the promoter sequences of crcZ, crcY, PP2810, oprD and PP3420 containing the CbrB binding sites. Grey boxes denote putative CbrB binding subsites in direct (type F) or reverse orientation (type R). In lowercase letters those bases that differ with the consensus sequence, which is denoted at the top.