Figure 1.
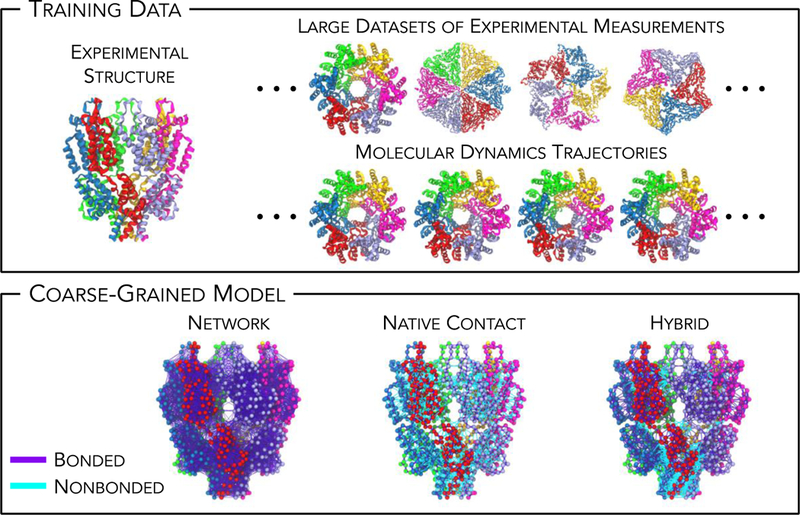
Different avenues of approach for model development of highly coarse-grained (CG) macromolecules, which are broadly classified into structure-based, knowledge-based, and dynamics-based strategies. Here, the capsid and spacer peptide 1 (CA-SP1) domains of the human immunodeficiency virus (HIV) protein is used as a representative example. (Top) Each model class relies on different training datasets and methods; for example, an experimental structure (PDB 5L93), an extended dataset of experimental measurements (PDB 5L93, 3ZX8, 3J37 and 6BVF), or statistics from an all-atom molecular dynamics trajectory (initial structures from PDB 5L93), respectively. (Bottom) Resultant models can be classified as network-based and native-contact-based models (or hybrids thereof); bonded (nonbonded) interactions are depicted by solid purple (cyan) lines while CG sites (per residue) are depicted as spheres.
