Figure 2. Network analyses of enzyme motions identify critical residues for allostery.
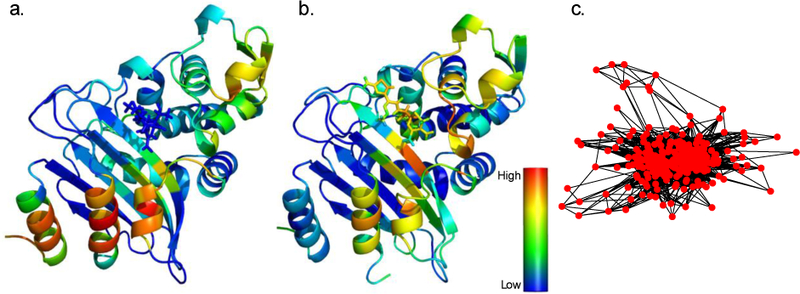
Rendered here are network centrality scores for the CTX-M9 beta lactamase based on positional mutual information in molecular dynamics simulations [50]. Coloring in panel (a) is based on network eigencentrality of the CTX-M9:cefotaxime acylenzyme complex, while coloring in panel (b) is based on the same complex with meropenem. The residue-residue interaction graph used to calculate (a) is rendered in panel (c). Network-based analyses have identified residues predicted to be either closely coupled to active site residues or broadly critical to beta-lactamase structure and dynamics. Edges in these networks can derive from both structure-based and dynamics-based methods; this rendering uses dynamics-based methods (symmetric uncertainty calculated from sitional mutual information). The symmetric uncertainty matrix is thresholded at 0.01 for graph rendering, although centrality is calculated on the full graph. Several important allosteric mutants score highly using this analysis, but such combinations of positional mutual information and network centrality await systematic experimental confirmation.
