Figure 3. Proposed coupling of large-scale molecular dynamics, QM/MM, and statistical learning for insight into allostery.
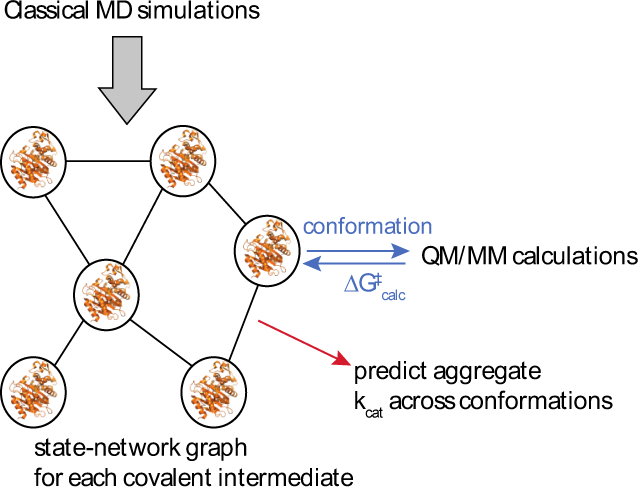
In this scheme, classical molecular dynamics simulations are used to construct a state network for each covalent intermediate in beta-lactamase hydrolysis. QM/MM calculations on conformations sampled from this state network are used to predict ∆G‡ for each conformation. These predictions are used both to guide lumping/splitting of the state network (to maintain uniform ∆G‡ within a state) and to calculate aggregate kcat across the states. This process could either be repeated across multiple enzyme mutants for retrospective analysis or used prospectively to guide targeted mutagenesis.
