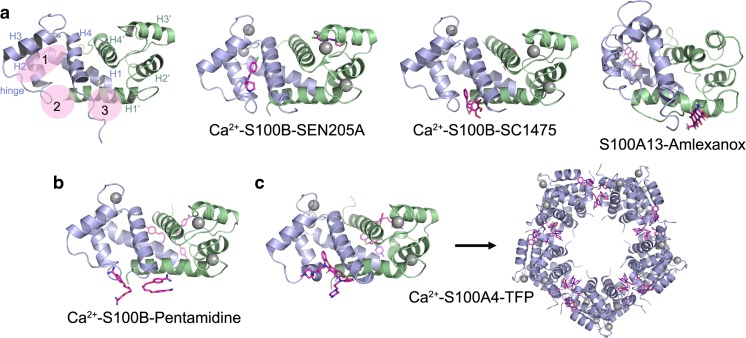
Fig. 3.
S100 protein-inhibitor complexes. a Ribbon diagram of Ca2+-S100B showing the general locations of the three binding sites that can accommodate small molecules and representative structures showing small molecules bound to each site. The individual S100 subunits are shown in light blue and green, the Ca2+ ions are shown as gray spheres and the inhibitors as pink sticks. Site 1: S100B-SEN205A (PDB 3HCM)—involves residues from the hinge and helices 2 and 3. Site 2: S100B-SC1475 (3,4-dimethoxydalbergione) (PDB 4PE4)—involves residues from the hinge and helix 4. Site 3: S100A13-amlexanox (PDB 2KOT)—involves residues from the C-terminal loop and helix 1. b Structures of Ca2+–S100B-pentamidine (PDB 3CR4) and c Ca2+–S100A4-trifluoperazine (TFP) (PDB 3KO0) showing two inhibitor molecules bound per S100 subunit and occupation of both sites 2 and 3. TFP binding induces the assembly of five Ca2+–S100A4-TFP dimers into a pentameric ring