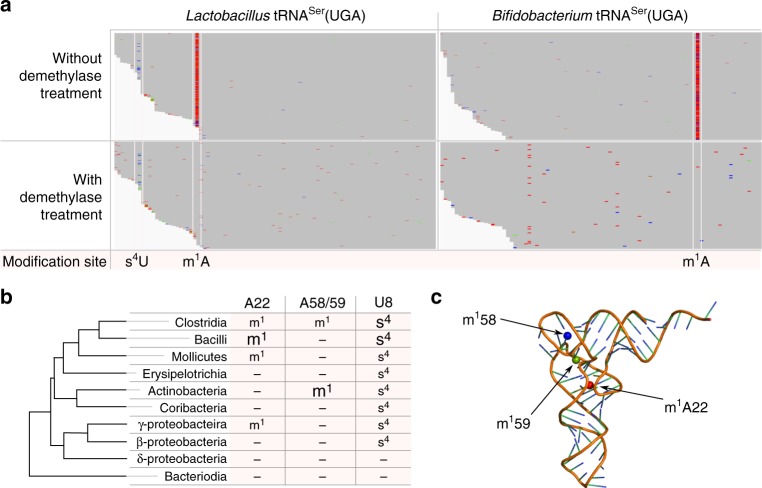
Fig. 5.
Taxonomic differences of modification sites. a Examples of aligning tRNA sequencing reads to two seed sequences of tRNASer(UGA) from Lactobacillus, class Bacilli, and Bifidobacterium, class Actinobacteria without and with demethylase treatment. Modification sites identified (s4U and m1A) are highlighted between the white lines. b m1A22, m1A58/59, and s4U8 identified in the abundant bacterial classes from Fig. 3c in the context of their phylogenetic relationship. Large fonts indicate bacterial classes in which the majority of the modifications are found (m1A22 in Clostridia and Bacilli, m1A58/59 in Actinobacteria, and s4U8 in Clostridia and Bacilli). c Proximal location of the m1A22 (red), m158 (blue), and m159 (green) modifications in a tRNA three-dimensional structure