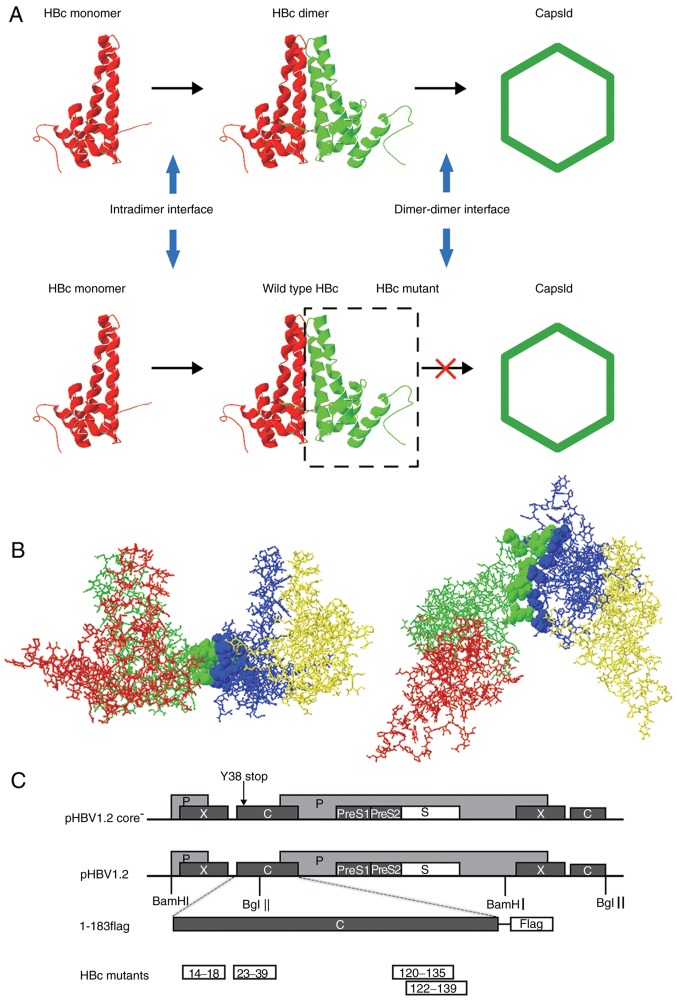
Figure 1.
HBV capsid assembly and dimer-dimer interface. (A) Capsid assembly procedure. Upper panel, two α-helical HBc monomers form a dimer via the intradimer interface, and numerous dimers subsequently form an icosahedral capsid via the dimer-dimer interface. Lower panel, HBc with mutations at the dimer-dimer interface may interact with WT HBc monomer via the intradimer interface to form an aberrant dimer, but disrupts the capsid structure or function. (B) Orthogonal perspectives of HBc (1QGT, Protein Data Bank) viewed normal to the local 2-fold axis (right) and along the 2-fold axis from the outside of the HBc (left); 4 HBc monomer units are indicated by four different colors (red, green, blue and yellow). Residues at the dimer-dimer interface are presented as ribbons, indicated in green (14E, 18F, 120V, 124V, 127R, 129P, 132Y, 134P and 135P) and blue (23F, 25P, 29D, 37L, 39R, 122F and 139I), respectively. (C) Map of plasmids. For the expression of the HBV genome, 1.2 units of the HBV genome was inserted at the 3′ end of the lac promoter in pUC18 to construct the pHBV1.2 plasmid. The open reading frames of the viral C, P, E and X genes are indicated by dark grey boxes. The C gene of pHBV1.2core− contained a point mutation of a stop codon at codon 38 (arrow). For the expression of WT and mutant HBc, the pcDNA3.1 vector was used. A FLAG tag was fused to the C-terminus of HBc. Each HBc mutant is indicated by a white box. HBV, hepatitis B virus; HBc, HBV core protein; WT, wild-type.