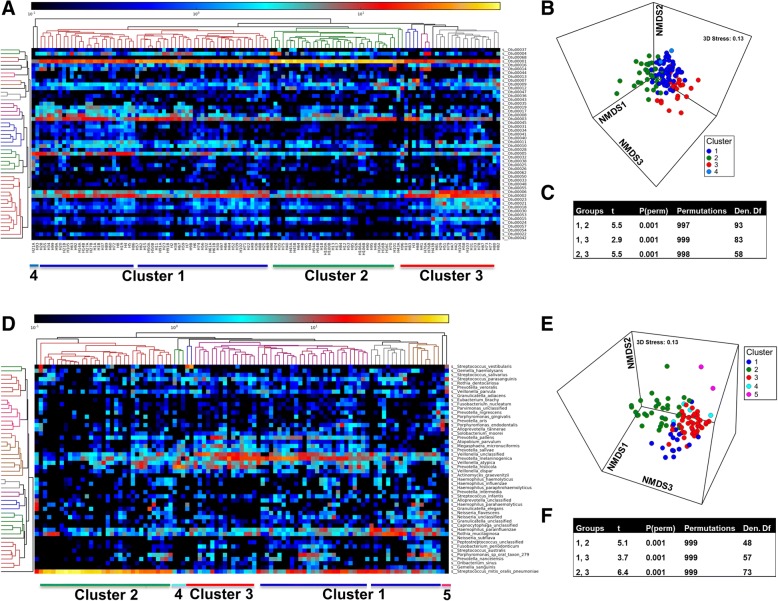
Fig. 1.
The esophageal microbiome clusters into different community types (esotypes). a Heatmap of relative abundances of the top 50 OTUs generated through 16S rRNA amplicon sequencing were used for a hierarchical cluster analysis (HCA) within MetaPhlan2. Dark blue to yellow correspond to 0.1–100% abundance. All available samples (n = 122) were utilized in this analysis. Taxonomy of OTUs is provided in Additional file 1.1. b Non-metric multidimensional scaling (nMDS) plot of Bray–Curtis resemblance generated from square root-transformed OTU relative abundances (all OTUs). OTU relative abundances were generated from 16S rRNA amplicon sequencing. Clusters from the HCA (panel a) were overlayed onto the nMDS plot. c PERMANOVA across HCA clusters of Bray–Curtis resemblance generated from square root-transformed OTU relative abundances. Test of the homogeneity of multivariate dispersions within groups at OTU level using PERMDISP showed no differences across clusters. ANOSIM generated a sample statistic of 0.42 and P = 0.001. d Relative abundances of the top 50 species generated through shotgun sequencing and MetaPhlan2 analysis were used for HCA. HCA was performed in MetaPhlan2. e nMDS plot of Bray–Curtis resemblance generated from square root-transformed species relative abundances (shotgun). All available shotgun samples were utilized in this analysis. Clusters from HCA of shotgun data (MetaPhlan2) were overlayed onto the nMDS plot. f PERMANOVA across shotgun HCA clusters of Bray–Curtis resemblance generated from square root-transformed species relative abundances. ANOSIM at the species level generated a sample statistic of 0.54 and P = 0.001