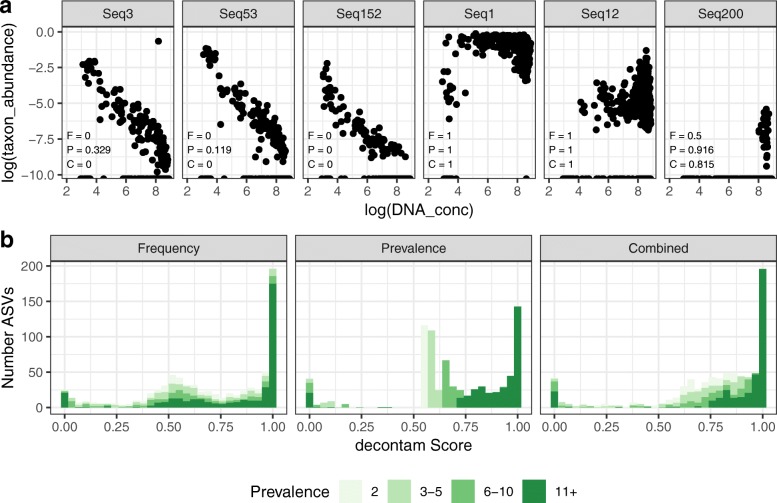
Fig. 2.
Frequency patterns and decontam scores of microbial sequences from an oral 16S rRNA gene dataset. a Frequency patterns of six sequences from a 16S rRNA gene study of human oral mucosal microbial communities. The frequencies of sequence variants Seq3, Seq152, and Seq53 vary inversely with sample DNA, a characteristic of contaminants. The frequencies of Seq1, Seq12, and Seq200 are independent of sample DNA concentration, a characteristic of genuine sample sequences. Scores for each of the six sequences were computed by the frequency (F), prevalence (P), and combined (C) methods as implemented in the isContaminant function in the decontam R package, and are indicated in the bottom left of each panel. b Scores for each amplicon sequence variant (ASV) present in two or more samples were computed as in a. The histogram of scores is shown, with color intensity depending on the number of samples (or prevalence) in which each ASV was present