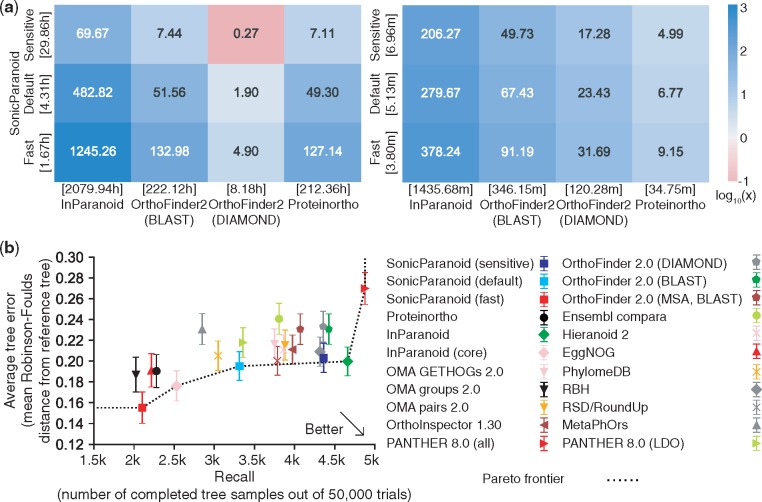
Fig. 1.
Speed and accuracy of SonicParanoid. (a) Speed-up of SonicParanoid (in the fast, default and sensitive modes) in relation to InParanoid, OrthoFinder2 (on BLASTP and DIAMOND) and Proteinortho on the complete 2011 version of the QfO dataset (left: complete execution, right: orthology inference step only). The numbers on the tiles and their colors indicate the speed-up folds. The numbers in the square brackets represent the execution time in hours (left) or minutes (right). Eight processors were used for each tool. (b) Accuracy of SonicParanoid and other 13 orthology inference tools assessed by the QfO benchmark (generalized species tree discordance test at the last universal common ancestor level). The x-axis represents the numbers of completed tree samples out of 50 000 trials (larger is better), while the y-axis represents the average tree error (smaller is better)