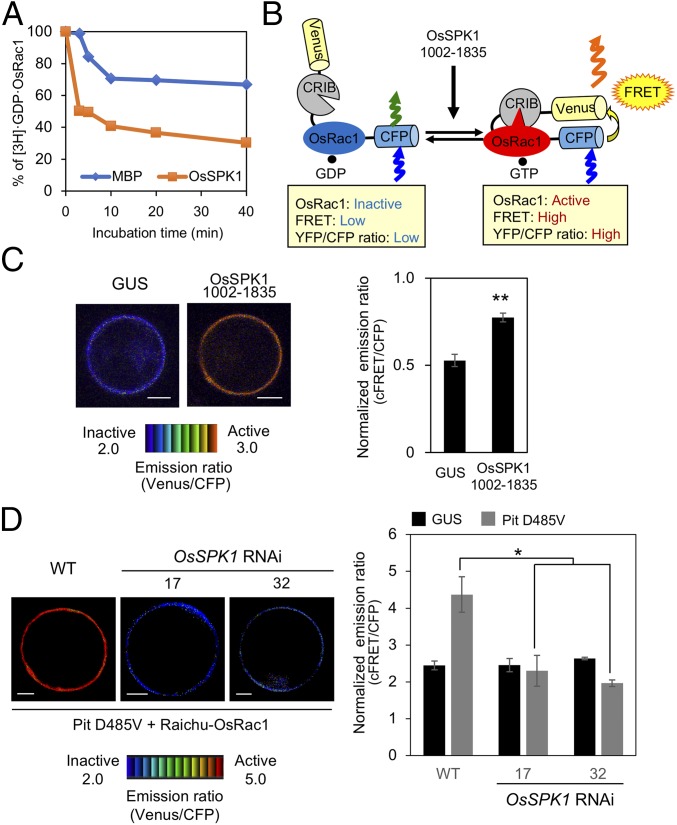
Fig. 4.
OsSPK1 functions as a GEF protein toward OsRac1. (A) In vitro GEF assay of OsSPK1. In vitro GEF activity of OsSPK1 was detected by the dissociation of [3H]-labeled GDP from OsRac1. MBP was a negative control. (B) Schematic overview of the Raichu-OsRac1 sensor used to monitor OsRac1 activation in vivo. (C) Detection of in vivo OsRac1 activation by OsSPK1 using Raichu-OsRac1 FRET. Emission ratio images (Left) of confocal laser-scanning micrographs of rice protoplasts coexpressing Raichu-OsRac1 and OsSPK1 (amino acids 1002–1835) or control GUS. The color scale from blue to red represents low to high levels of OsRac1 activation. (Scale bars, 5 μm.) Statistical analysis (Right) of OsRac1 activation using Raichu-OsRac1 with normalized emission ratios of Venus to CFP. Bars are SE (**P < 0.01, n = 60). (D) Effect of OsSPK1 RNAi on Pit D485V-induced OsRac1 activation. Emission ratio images (Left) of confocal laser-scanning micrographs of OsSPK1 RNAi and WT rice protoplasts coexpressing Raichu-OsRac1 and Pit D485V or control GUS. The color scale from blue to red represents low to high levels of OsRac1 activation. (Scale bars, 5 μm.) Statistical analysis (Right) of OsRac1 activation using Raichu-OsRac1 with normalized emission ratios of Venus to CFP. Bars are SE (*P < 0.05, n = 60).