Significance
Mucosa-associated invariant T (MAIT) cells are unconventional innate-like T cells recognizing microbial riboflavin metabolites presented by the monomorphic MR1 molecule. Here, we show that the CD8+CD4− and CD8−CD4− subpopulations of human MAIT cells represent transcriptionally and phenotypically discrete subsets with distinct functional profiles. Furthermore, T cell receptor repertoire analysis, as well as MAIT cell data based on human fetal tissues, umbilical cord blood, and culture systems indicate that the CD8−CD4− subset may derive from the main CD8+CD4− MAIT cell pool. Thus, MAIT cells, a major antimicrobial effector T cell population in humans, segregate into two functionally distinct but developmentally related subsets separated by the expression of CD8. This functional difference may have significant implications in infectious and inflammatory diseases.
Keywords: MAIT cells, MR1, CD8, apoptosis, functional heterogeneity
Abstract
Mucosa-associated invariant T (MAIT) cells are unconventional innate-like T cells that recognize microbial riboflavin metabolites presented by the MHC class I-like protein MR1. Human MAIT cells predominantly express the CD8α coreceptor (CD8+), with a smaller subset lacking both CD4 and CD8 (double-negative, DN). However, it is unclear if these two MAIT cell subpopulations distinguished by CD8α represent functionally distinct subsets. Here, we show that the two MAIT cell subsets express divergent transcriptional programs and distinct patterns of classic T cell transcription factors. Furthermore, CD8+ MAIT cells have higher levels of receptors for IL-12 and IL-18, as well as of the activating receptors CD2, CD9, and NKG2D, and display superior functionality following stimulation with riboflavin-autotrophic as well as riboflavin-auxotrophic bacterial strains. DN MAIT cells display higher RORγt/T-bet ratio, and express less IFN-γ and more IL-17. Furthermore, the DN subset displays enrichment of an apoptosis gene signature and higher propensity for activation-induced apoptosis. During development in human fetal tissues, DN MAIT cells are more mature and accumulate over gestational time with reciprocal contraction of the CD8+ subset. Analysis of the T cell receptor repertoire reveals higher diversity in CD8+ MAIT cells than in DN MAIT cells. Finally, chronic T cell receptor stimulation of CD8+ MAIT cells in an in vitro culture system supports the accumulation and maintenance of the DN subpopulation. These findings define human CD8+ and DN MAIT cells as functionally distinct subsets and indicate a derivative developmental relationship.
Human mucosa-associated invariant T (MAIT) cells are unconventional T cells defined by their semi-invariant T cell receptor (TCR) containing the invariant Vα7.2 segment coupled with Jα12, 20, or 33, and limited β-chain diversity (1–5). MAIT cells are highly abundant in peripheral blood, mucosal tissues, and the liver (5–7). They express high levels of the NK cell-associated receptors CD161 and IL-18Rα, and tissue-homing chemokine receptors including CCR5, CCR6, CXCR6, as well as α4β7 (5–9). MAIT cells recognize antigens derived from the riboflavin biosynthetic pathway produced by a wide range of microbes and presented by the MHC class-I related (MR1) protein (10, 11). Following activation, MAIT cells rapidly produce cytokines—including IFN-γ, TNF, IL-17, and IL-22 (6, 9, 12, 13)—and mediate cytolytic activity against infected cells (14–16), leading to the control of various bacterial infections in the settings of human infectious diseases and in animal models (reviewed in ref. 17). Innate cytokines, such as IL-12 and IL-18, can activate some functions in MAIT cells in an MR1-independent fashion (18, 19), and enhance MAIT cells’ TCR-dependent activation (20, 21). MR1-independent responses are likely important for the involvement of MAIT cells in viral infections (22–28), and in diseases driven by aberrant cytokine release caused by bacterial exotoxins (29, 30).
MAIT cells develop in the thymus, positively selected by MR1-expressing CD4+CD8+ (double-positive, DP) thymocytes (5, 7, 31, 32). In the thymus of both mice and humans, three stages of MAIT cells have been identified: stage 1 cells, predominantly DP and CD4+CD8− (CD4+); stage 2 cells, predominantly DP, CD4+, and CD4−CD8+ (CD8+); and stage 3 cells, predominantly CD8+ and CD4−CD8− [double negative (DN)], and less abundant than the other two stages (33). In the circulation of healthy adults, MAIT cells are predominantly CD8+ with a smaller DN subset (4, 7, 33). Thus, the DN MAIT cells are relatively rare in the thymus but more abundant in the peripheral blood of healthy adults. There is currently a paucity of information concerning the immunobiology of the CD8+ and DN MAIT cell subsets. Here, we investigated in detail the phenotypic, transcriptional, and functional differences between CD8+ and DN MAIT cells using human samples from peripheral blood, mucosal tissues, and fetal tissues. Our data indicate that the CD8+ and DN MAIT cells represent functionally distinct subsets, and suggest that DN MAIT cells may be derived from the main CD8+ MAIT cell pool.
Results
CD8+ and DN MAIT Cells Are Consistently MR1-Restricted.
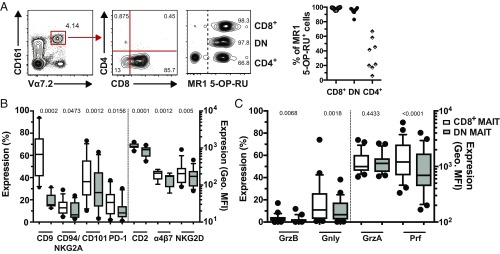
We initially investigated the ability of peripheral blood CD8+ and DN CD161hiVα7.2+ MAIT cells to bind to their MR1 restriction element (Fig. 1A and SI Appendix, Fig. S1A). The vast majority of CD8+ and DN MAIT cells stained with the MR1 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) tetramer [median (interquartile range, IQR) percentage of MR1 5-OP-RU+ cells: 98.7% (97.15–99.25) and 96.15% (93.48–97.58) for CD8+ and DN MAIT cells, respectively] (Fig. 1A), and did not stain with the MR1 6-formyl pterin- (6-FP) negative control tetramer (SI Appendix, Fig. S1B), as recently reported (34). In contrast, only one-third of the CD4+ CD161hiVα7.2+ T cells stained with the MR1 5-OP-RU tetramer (Fig. 1A) [median (IQR): 31.55% (17.9–55.03)], as previously reported (3). The CD161−Vα7.2+ T cells contained a negligible MAIT cell population (SI Appendix, Fig. S1C). Thus, the vast majority of peripheral blood CD8+ and DN CD161hiVα7.2+ MAIT cells are MR1-restricted.
Fig. 1.
Human CD8+ MAIT cells display higher expression of coactivating receptors and cytotoxic molecules. (A) Representative flow cytometry identification of CD161hiVα7.2+ MAIT cells, CD4 and CD8 staining within the MAIT cell population, and MR1 5-OP-RU tetramer staining within CD8+, DN, and CD4+ CD161hiVα7.2+ MAIT cells (Left). Percentage of MR1 5-OP-RU+ cells within the CD161hiVα7.2+ CD8+, DN, and CD4+ MAIT cells (Right). (B) The expression of the coactivating receptors CD2 and CD9, as well as of CD101, α4β7, PD-1, CD94, NKG2A, and NKG2D on CD8+ and DN MAIT cells. (C) The expression of cytolytic proteins in CD8+ and DN MAIT cells. Data are from 8 to 10 donors for MR1 5-OP-RU tetramer staining (A), 10 donors for surface marker expression (B), and 25 donors for cytolytic protein expression (C). Box-and-whisker plots show the median, the 10th and 90th percentiles, and the IQR. The paired t test was used to detect significance between paired samples, except for PD-1, NKG2D, Gnly, and Prf, where the Wilcoxon’s signed-rank test was used.
CD8+ MAIT Cells Express Higher Levels of Coactivating Receptors and Cytolytic Effector Molecules than DN MAIT Cells.
To investigate the surface immunoreceptor profile of CD8+ and DN MAIT cells, resting peripheral blood mononuclear cells (PBMCs) from healthy individuals were prestained for CD3, CD161, and Vα7.2, and then screened for 332 surface proteins by flow cytometry, as previously described (8). The two MAIT cell subsets displayed a high degree of similarity in their overall surface immunoproteome (SI Appendix, Table S1). However, close inspection of this dataset revealed clear differences between the subsets, because CD8+ MAIT cells expressed CD2, CD9, as well as the CD8+ regulatory T cell-associated protein CD101, at higher levels than their DN counterparts (SI Appendix, Table S1). Repeat experiments confirmed higher expression of CD2, CD9, and CD101 on CD8+ MAIT cells (all P < 0.01) (Fig. 1B and SI Appendix, Fig. S1D). Moreover, the expression of programmed death-1 receptor (PD-1) and the gut homing integrin α4β7 was higher in CD8+ MAIT cells (both P < 0.05) (Fig. 1B and SI Appendix, Fig. S1D). The surface immunoproteome dataset indicated a difference in the expression of CD94 (SI Appendix, Table S1), and the NK cell receptor defined by coexpression of CD94 and NKG2A was expressed at higher levels by CD8+ MAIT cells (P = 0.047) (Fig. 1B and SI Appendix, Fig. S1D). CD8+ MAIT cells also expressed higher levels of NKG2D, another member of the NKG2 family (P = 0.005) (Fig. 1B and SI Appendix, Fig. S1D). Furthermore, CD8+ MAIT cells had significantly higher granzyme (Grz) B, granulysin (Gnly), and perforin (Prf) levels than DN MAIT cells (all P < 0.01) (Fig. 1C and SI Appendix, Fig. S1D). GrzA was expressed at similar levels between the two subsets (Fig. 1C and SI Appendix, Fig. S1D). Taken together, these data indicate that peripheral blood CD8+ MAIT cells display higher baseline expression of coactivating receptors and cytotoxic effector molecules than DN MAIT cells.
CD8+ and DN MAIT Cell Subpopulations Display Distinct Transcriptional Profiles.
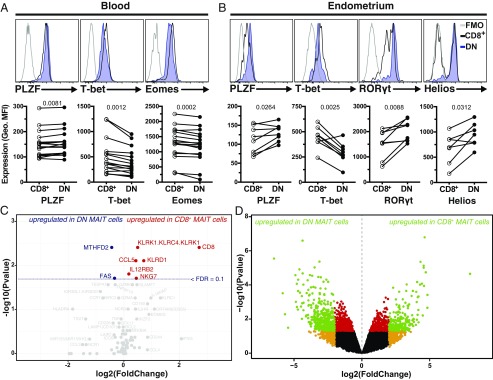
Next, we investigated the intracellular expression of several transcription factors in CD8+ and DN MAIT cells from peripheral blood (16), including promyelocytic leukemia zinc finger protein (PLZF), T box transcription factor 21 (TBX21 or T-bet), eomesodermin (Eomes), retinoid-related orphan receptor (ROR) γt, and Helios (Fig. 2 A and B). Circulating CD8+ MAIT cells expressed significantly lower levels of PLZF and higher levels of T-bet and Eomes compared with the DN subset (all P < 0.01) (Fig. 2A). Expression of RORγt and Helios was not significantly different (P = 0.12 and P = 0.17, respectively) (SI Appendix, Fig. S2A). Interestingly, the modest differences observed in peripheral blood MAIT cells were more pronounced in mucosal MAIT cells isolated from the endometrium (Fig. 2B) (12). Here, the expression levels of PLZF, RORγt, and Helios were significantly higher in DN MAIT cells, whereas T-bet expression was higher in the CD8+ subset (all P < 0.05) (Fig. 2B). There was no difference in Eomes expression between endometrial CD8+ and DN MAIT cells (P = 0.43) (SI Appendix, Fig. S2B). Together, these data indicate that CD8+ and DN MAIT cells display distinct transcription factor expression patterns, particularly in genital mucosal tissue.
Fig. 2.
CD8+ and DN MAIT cells express different levels of critical transcription factors. Representative example and geometric MFI of the staining intensity of (A) PLZF, T-bet, and Eomes in peripheral blood CD8+ and DN MAIT cells, and (B) PLZF, T-bet, RORγt, and Helios in endometrium-derived CD8+ and DN MAIT cells. The fluorescence minus one (FMO) control is included as control. (C and D) Volcano plots showing fold-changes in the expression of individual genes in CD8+ MAIT cells compared with DN MAIT cells as a function of their P values, as determined by Fluidigm Biomark (C) or RNAseq (D). Dotted line represents filtering criteria of FDR = 0.1 (C). Filtering criteria for green, red, and orange data points are P < 0.05 and absolute log2(fold-change) > 2; P < 0.05; absolute log2(fold-change) > 2, respectively (D). Data are from 16 (A), 8 (B), 9 (C), and 4 (D) donors (seven donors for Helios in B). Lines in the graphs represent individual donors. The paired t test was used to detect significant differences between paired samples, except for PLZF (A) and Helios (B), where the Wilcoxon’s signed-rank test was used.
To assess if the marked differences between CD8+ and DN MAIT cells were also reflected at the gene expression level, their targeted transcriptomic signatures were explored in a selected set of 85 genes of interest using the Fluidigm Biomark platform (SI Appendix, Table S2). First, CD8α gene expression was considerably higher in the CD8+ MAIT cells than in the DN MAIT cells (Fig. 2C), demonstrating that the absence of CD8 on the cell surface of DN MAIT cells was not a mere surface down-regulation. Second, killer cell lectin-like receptor subfamily D1 (KLRD1, or CD94) and KLRK1 (NKG2D) genes were more highly expressed in CD8+ MAIT cells (Fig. 2C), in line with the immunoproteome dataset (Fig. 1B and SI Appendix, Table S1). In addition, the expression of genes encoding the NK cell granule protein 7 (Nkg7), the C-C motif chemokine ligand 5 (Ccl5), and the IL-12 receptor β2 subunit (Il12rb2) was higher in CD8+ MAIT cells (Fig. 2C). Third, compared with DN MAIT cells, the CD8+ MAIT cell subset displayed modestly lower expression of genes encoding the first apoptosis signal receptor (Fas), and the bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD2) involved in folate metabolism (Fig. 2C).
Based on the initial indications of differential gene-expression patterns, CD8+ and DN peripheral blood MAIT cell subsets were again sorted from new donors for whole-genome transcriptional analysis using RNA sequencing (RNAseq). We identified a core signature of 598 genes that were differentially expressed between CD8+ and DN MAIT cells, with 214 up-regulated in CD8+ MAIT cells, including the CD8α and CD8β genes, and 384 genes up-regulated in the DN MAIT cells (Fig. 2D and SI Appendix, Table S3). t-Distributed stochastic neighbor embedding (t-SNE) analysis supported that CD8+ and DN MAIT cells represent distinct populations based on their differential transcriptome pattern (SI Appendix, Fig. S2C). Altogether, these data demonstrate that peripheral blood CD8+ and DN MAIT cells have distinct transcriptional signatures.
Differential Functional Profile of the Two MAIT Cell Subsets.
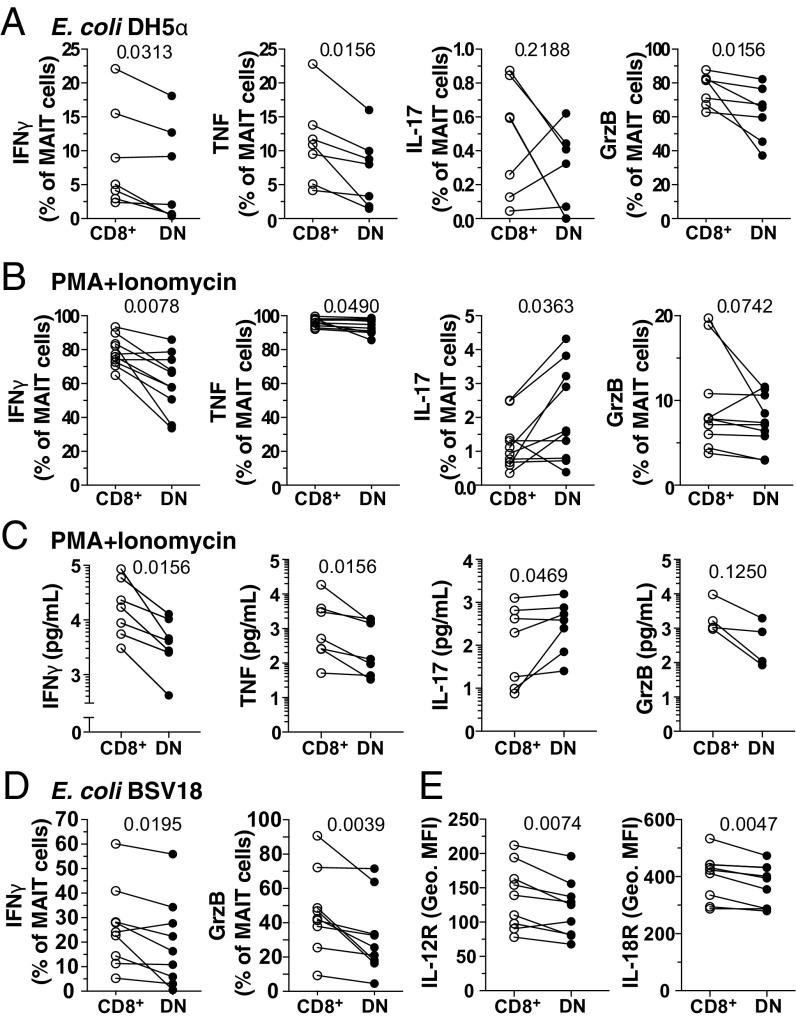
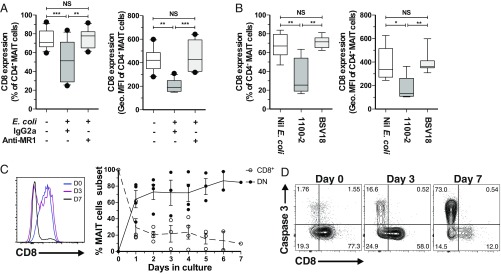
To explore whether these phenotypic and transcriptomic differences translate into differential functional capacity, their responsiveness to Escherichia coli and phorbol myristate acetate (PMA)/ionomycin in vitro stimulations was examined. Sorted CD8+ and DN MAIT cells were stimulated with autologous E. coli-fed monocytes or with PMA/ionomycin (35), and intracellularly stained for the cytokines IFN-γ, TNF, and IL-17, as well as for the cytolytic protein GrzB (Fig. 3 A and B and SI Appendix, Fig. S3A). CD8+ MAIT cells responded to both E. coli- and PMA/ionomycin-mediated stimulations with significantly higher production of IFN-γ and TNF than the DN MAIT cells (all P < 0.05) (Fig. 3 A and B). GrzB production was also higher in CD8+ MAIT cells upon E. coli-mediated stimulation (P = 0.0156) (Fig. 3 A and B). Levels of IFN-γ and TNF expression as determined by the geometric mean fluorescence intensity (MFI) by PMA/ionomycin-stimulated CD8+ MAIT cells were higher, as well as of GrzB expression by E. coli-stimulated CD8+ MAIT cells compared with their DN counterparts (SI Appendix, Fig. S3B). In contrast, DN MAIT cells expressed slightly higher levels of IL-17 than the CD8+ MAIT cells in response to PMA/ionomycin stimulation (P = 0.0363) (Fig. 3B). Both CD8+ and DN MAIT cells responded to E. coli in a predominantly MR1-dependent manner, as determined by MR1-blocking (SI Appendix, Fig. S3C). Quantification of these cytokines and cytolytic molecules in the supernatant of PMA/ionomycin-stimulated MAIT cells (Fig. 3C) revealed a similar pattern as the intracellular staining (Fig. 3B). Specifically, CD8+ MAIT cells produced significantly higher levels of IFN-γ and TNF, and lower levels of IL-17, compared with DN MAIT cells (Fig. 3C).
Fig. 3.
DN MAIT cells display less functional capacity following bacterial and mitogenic stimulation than CD8+ MAIT cells. Percentage of FACS-sorted CD8+ and DN MAIT cells expressing IFN-γ, TNF, IL-17, and GrzB after stimulation with (A) E. coli for 24 h (n = 7) and (B) PMA/ionomycin for 6 h (n = 10). (C) Concentration of IFN-γ, TNF, IL-17, and GrzB after stimulation with PMA/ionomycin for 6 h (n = 4–7). (D) Expression of IFN-γ and GrzB by MAIT cells within bulk PBMC culture in the presence of RibA− E. coli BSV18 (n = 9). (E) Expression levels (geometric MFI) of IL-12R and IL-18R in resting CD8+ and DN MAIT cells (n = 9). Lines in the graphs represent individual donors. The Wilcoxon’s signed-rank test was used to detect significant differences between paired samples, except for IFN-γ, TNF, and IL-17 in the PMA/ionomycin stimulation where the paired t test was used.
To determine if the functional differences between MAIT cell subsets were MR1-dependent, we utilized the RibA-deficient E. coli strain BSV18 unable to synthesize riboflavin (SI Appendix, Fig. S3D) (36, 37). We confirmed the lack of MR1-dependent stimulation of MAIT cells by the BSV18 strain, in contrast to its riboflavin biosynthesis-competent congenic strain 1100-2 (SI Appendix, Fig. S3 E and F). Because BSV18 did not induce significant CD8 down-regulation (SI Appendix, Fig. S3G), we measured cytokine production to this bacterial strain by CD8+ and DN MAIT cells after stimulation of PBMC cultures. CD8+ MAIT cells expressed significantly higher levels of IFN-γ and GrzB following BSV18-mediated stimulation (both P < 0.05) (Fig. 3D). There were no differences in the TNF and IL-17 levels (SI Appendix, Fig. S3H); however, these cytokines were expressed at much lower levels than usually observed, likely due to the lack of MR1-presented antigens (8). Because IL-12 and IL-18 production by antigen-presenting cells (APCs) is involved in MR1-independent activation of MAIT cells (19), we next determined the expression of IL-12R and IL-18R on MAIT cell subsets. The DN subset had significantly lower IL-12R and IL-18R levels (Fig. 3E), in line with the transcriptome data (Fig. 2C). Consistent with this finding, CD8+ MAIT cells produced slightly more IFN-γ and GrzB than DN MAIT cells following IL-12+IL-18 stimulation (SI Appendix, Fig. S3I). The lower responses of the DN subset following MR1-independent E. coli BSV18 stimulation may thus be partly caused by the lower response to IL-12 and IL-18. Taken together, these data indicate that peripheral blood CD8+ MAIT cells respond more strongly in terms of IFN-γ, TNF, and GrzB production to TCR-dependent and -independent, as well as to mitogen-mediated stimulations. This is consistent with their higher basal expression of IL-12R, IL-18R (Fig. 3E), coactivating receptors, and cytotoxic molecules (Fig. 1 B and C), and the classic effector transcription factors Eomes and T-bet (Fig. 2 A and B).
DN MAIT Cells Have Higher Levels of a Proapoptotic Signature than CD8+ MAIT Cells.
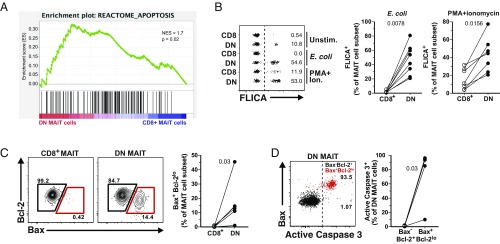
We next revisited the RNAseq data and performed gene set enrichment analysis (GSEA) of the transcripts differentially expressed between MAIT cell subsets. The GSEA analysis using the Reactome database revealed an enrichment of an apoptosis gene signature in DN MAIT cells (Fig. 4A). To extend these findings, we next evaluated the apoptosis propensity of the two MAIT cell subsets in vitro. Upon stimulation of FACS-sorted CD8+ and DN MAIT cells with E. coli-fed monocytes or PMA/ionomycin, DN MAIT cells were significantly more prone to apoptosis than CD8+ MAIT cells, as measured by the percentage of FLICA+ cells after stimulation (both P < 0.05) (Fig. 4B). Following E. coli-mediated stimulation, the increase in apoptosis was predominantly MR1-dependent in both subsets, as the percentage of FLICA+ MAIT cells decreased upon MR1 blocking (both P < 0.05) (SI Appendix, Fig. S4A).
Fig. 4.
DN MAIT cells are more prone to apoptosis than CD8+ MAIT cells. (A) Gene-set enrichment summary plot for CD8+ MAIT cell versus DN MAIT cell differentially expressed ranked genes and a defined set of genes associated with an apoptosis signature. (B) Representative example of FLICA expression in FACS-sorted CD8+ and DN MAIT cells in the absence of stimulation, or after E. coli- or PMA/ionomycin-mediated stimulations (Left). Summary data with the percentage of stimulated CD8+ and DN MAIT cells expressing FLICA (Right). (C) Representative FACS plots and the frequency of Bax and Bcl-2 expression in resting CD8+ and DN MAIT cells. (D) Representative FACS plot and expression of active caspase 3 in Bax+Bcl-2lo (red gate) DN MAIT cells. Data are from seven (A), eight (B; E. coli), seven (B; PMA/ionomycin), and six (C and D) donors. Lines in the graphs represent individual donors. The Wilcoxon’s signed-rank test was used to detect significant differences between paired samples. NES, normalized enrichment score.
We next investigated the baseline expression of the antiapoptotic protein Bcl-2 and the proapoptotic Bcl-2 associated X, apoptosis regulator (Bax). Resting DN MAIT cells had higher frequency of Bax+Bcl-2lo cells (P = 0.03) (Fig. 4C), and the Bax+Bcl-2lo phenotype was linked to profoundly higher levels of active caspase 3 in DN MAIT cells (P = 0.03) (Fig. 4D). Altogether, these data indicate that DN MAIT cells are more prone to apoptosis, consistent with their higher Bax/Bcl-2 ratio and PLZF expression. The former determines cell survival or death following an apoptotic stimulus, and the latter was previously associated with the higher apoptotic propensity of MAIT and invariant natural killer T (iNKT) cells (38, 39). In light of these results, we evaluated whether the lower functionality of DN MAIT cells (Fig. 3 A–D) was due to the higher proportion of apoptotic cells in this subset. When the functional readouts were assessed in nonapoptotic (DCM−FLICA−) MAIT cells, the nonapoptotic DN MAIT cells still produced less IFN-γ and TNF than the CD8+ MAIT cells (both P < 0.05) (SI Appendix, Fig. S4B).
DN MAIT Cells Display a More Mature Phenotype During Human Fetal Development and a Restricted TCR Repertoire in Adult Peripheral Blood.
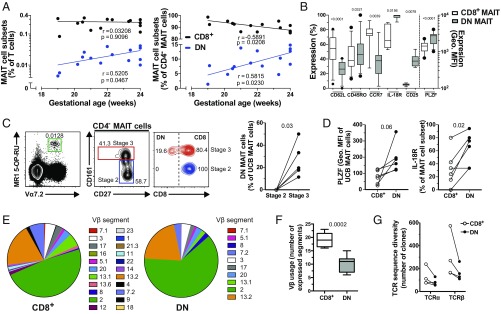
Given that DN MAIT cells in mice are detected predominantly during the later stages of thymic development (33), and that fetal human CD8αα MAIT cells may be derived from CD8αβ MAIT cells in a stepwise manner (13), we hypothesized that DN MAIT cells may derive from CD8+ MAIT cells in vivo. To examine this, we revisited our data on the CD8+ and DN MAIT cell subsets from second trimester human fetal spleens (13). The levels of DN MAIT cells in fetal spleens increased fivefold over gestational weeks 19–24, while the levels of CD8+ MAIT cells remained constant (Fig. 5A). To exclude contaminating non-MAIT cells within the CD4+ CD161hiVα7.2+ cell population, we gated on CD4− MAIT cells only and found that the size of the fetal splenic DN MAIT cell compartment similarly increased, with a respective decrease of CD8+ MAIT cell levels over developmental time (Fig. 5A). DN MAIT cells in the fetal spleen expressed higher levels of the activation marker CD25, as well as of PLZF (both P < 0.01) (Fig. 5B and SI Appendix, Fig. S5A). This pattern is consistent with the higher PLZF expression in DN MAIT cells from adult peripheral blood and endometrial tissues (Fig. 2 A and B). Fetal DN MAIT cells expressed lower levels of CD62L and CCR7, as well as higher levels of CD45RO and IL-18R (all P < 0.05) (Fig. 5B and SI Appendix, Fig. S5A), indicative of a more mature phenotype.
Fig. 5.
Fetal DN MAIT cells express a more mature phenotype and adult DN MAIT cells express a more restricted TCR repertoire than their CD8+ counterparts. (A) Correlations between the fetal gestational age and the levels of fetal splenic CD8+ and DN MAIT cells as a proportion of T cells (Left) and of CD4− MAIT cells (Right). (B) Expression of CD62L, CD45RO, CCR7, IL-18Rα, CD25, and PLZF in fetal splenic CD8+ and DN MAIT cells. Percentage of cells expressing each of these markers, except for PLZF for which the geometric MFI of the staining is shown. (C) Representative example of the identification of stage 2 and stage 3 MAIT cells from UCB and their CD8 expression, and the percentage of DN MAIT cells within each developmental stage. (D) PLZF and IL-18R expression by CD8+ and DN UCB MAIT cells. (E) Median percentage of adult circulating CD8+ and DN MAIT cells expressing each TCR Vβ segment. (F) Number of TCR Vβ segments expressed by adult circulating CD8+ and DN MAIT cells. (G) TCR-α and TCR-β sequence diversity of CD8+ and DN MAIT cells as determined by RNA sequencing. Data are from 15 donors (A); 11 (B; CD62L and CD45RO), 9 (B; CCR7), 7 (B; IL-18R), 8 (B; CD25), and 12 (B; PLZF) donors (B); 6 donors (C and D); 16–19 (E; CD8+ MAIT) and 14–17 (E; DN MAIT) donors (E); 9 (F; CD8+ MAIT) and 7 (F; DN MAIT) donors (F); and 4 donors (G). The box-and-whisker plots show the median, the 10th and 90th percentiles, and the IQR. Correlations were calculated using the Spearman’s test. The Wilcoxon’s signed-rank test was used to detect differences between paired samples for IL-18R, CCR7, CD25 (B) and in C, and the paired t test was used for the remainder (B and D). The unpaired t test was used to detect significant differences between unpaired samples (F).
To study this in more detail, MAIT cells were next characterized using the MR1 5-OP-RU tetramer in term umbilical cord blood (UCB). Among UCB MR1 5-OP-RU+ Vα7.2+ MAIT cells, the DN subset was only present in the more mature stage 3 CD27+CD161hi MAIT cell population, but not among the less mature stage 2 CD27+CD161−/dim cells (Fig. 5C) (33). Expression of PLZF and IL-18R primarily occurred in the CD161hi (stage 3) UCB MAIT cells consistent with a mature phenotype (13, 33) (SI Appendix, Fig. S5B). Accordingly, UCB DN MAIT cells expressed higher PLZF and IL-18R levels than their CD8+ counterparts (Fig. 5D). Altogether, these data indicate that DN MAIT cells increase in proportion during gestation, occur mostly in the stage 3 MAIT cell population, and are more mature than fetal CD8+ MAIT cells.
We next investigated the repertoire of TCR Vβ segments expressed by adult peripheral blood CD8+ and DN MAIT cells (Fig. 5E). CD8+ MAIT cells expressed a more diverse Vβ repertoire than DN MAIT cells (P = 0.0002) [median (IQR) of the number of Vβ segments: 19.0 (16.5–21.5) and 11.0 (7.0–12.0) by CD8+ and DN MAIT cells, respectively] (Fig. 5 E and F). Importantly, the vast majority of Vβ segments expressed by DN MAIT cells was also expressed by the CD8+ subset, but not vice versa (Fig. 5E). This pattern was consistent in each donor individually (SI Appendix, Fig. S5C). Finally, we analyzed the TCR transcripts within the RNAseq data from the sorted CD8+ and DN MAIT cell subsets (Fig. 5G). In the four donors analyzed, the CD8+ MAIT cell subset had a broader TCR diversity as assessed by the number of discrete TCR-α– and -β–chain clones detected (Fig. 5G). Both TCR chains displayed an overlap in clone sequences between CD8+ and DN MAIT subsets (SI Appendix, Fig. S5D), illustrating that there is a shared repertoire between the two subsets. Taken together, these observations support the notion that the DN MAIT cell subset may derive from the larger and more diverse CD8+ MAIT cell subpopulation.
Derivation of DN MAIT Cells from CD8+ MAIT Cells in Vitro.
The observation of CD8 down-regulation on MAIT cells following MR1-dependent stimulation with E. coli (SI Appendix, Fig. S6A), prompted us to investigate if activation may trigger the generation of the DN MAIT cell subset. MR1-blocking during stimulation with E. coli DH5α prevented CD8 down-regulation (Fig. 6A). Further experiments using the riboflavin autotroph E. coli 1100-2 also showed strong CD8 down-regulation, which did not occur when MAIT cells were stimulated with its riboflavin auxotroph congenic strain BSV18 (Fig. 6B and SI Appendix, Fig. S3G). In addition, stimulation with IL-12 and IL-18 did not lead to CD8 down-regulation (SI Appendix, Fig. S6B). Taken together, these data suggest that CD8 down-regulation by MAIT cells requires MR1 presentation of metabolite antigens to the MAIT cell TCR.
Fig. 6.
DN MAIT cells derived from CD8+ MAIT cells in vitro. (A) CD8 expression as percentage (Left) and geometric MFI (Right) in unstimulated and E. coli DH5α-stimulated MAIT cells in the presence of anti-MR1 mAb or isotype control (n = 15). (B) CD8 expression as percentage (Left) and geometric MFI (Right) in unstimulated and riboflavin autotroph E. coli 1100-2− or riboflavin auxotroph E. coli BSV18-stimulated MAIT cells (n = 11). (C) Representative histogram of CD8 expression (Left) and percentage of CD8+ and DN MAIT cell subsets (Right), and (D) representative FACS plots of active caspase 3 expression, during in vitro culture of FACS-sorted MAIT cells in the presence of immobilized Vα7.2 and CD28 mAb. The box-and-whisker plots show the median, the 10th and 90th percentiles, and the IQR. The lines and error bars represent mean and SE. The Friedman test followed by Dunn’s post hoc test was used to detect significant differences between multiple, paired samples (A and B). *P < 0.05, **P < 0.01, ***P < 0.001. NS, not significant.
Next, we examined if DN MAIT cells can be derived from CD8+ MAIT cells in vitro. To mimic MR1-restricted antigen presentation, FACS-sorted MR1 5-OP-RU+ Vα7.2+ CD161hi CD8+ MAIT cells were cultured in an APC-free system in the presence of immobilized Vα7.2 and CD28 mAbs. The down-regulation of CD8 and the appearance of DN MAIT cells were rapid and persisted throughout the 7-d culture (Fig. 6C). This was in contrast to TCR Vα7.2 down-regulation, which was transient (SI Appendix, Fig. S6C), indicating that CD8 down-regulation was not directly linked to the internalization of the TCR complex. The expansion of DN MAIT cells was independent of proliferation (SI Appendix, Fig. S6C), and appeared to be followed by apoptosis of the DN MAIT cell subset, as measured by active caspase 3 expression (Fig. 6D and SI Appendix, Fig. S6D). MAIT cells maintaining CD8 expression had negligible active caspase 3 expression (Fig. 6D and SI Appendix, Fig. S6D), consistent with our observation that CD8+ MAIT cells were far less prone to apoptosis (Fig. 4). Applying a similar experimental approach to FACS-sorted DN MAIT cells did not lead to the appearance of CD8+ MAIT cells in cultures (SI Appendix, Fig. S6E). Taken together, our data support a model where DN MAIT cells can be derived from CD8+ MAIT cells as a consequence of TCR-mediated activation.
Discussion
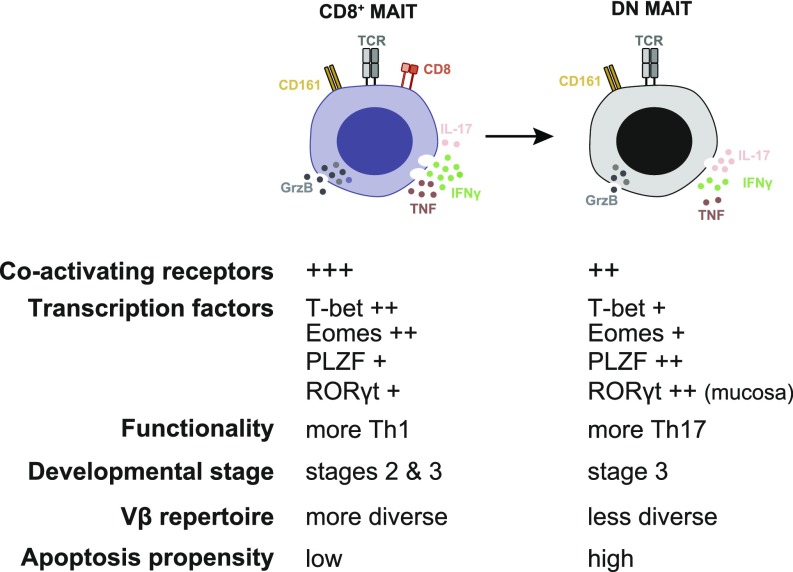
MAIT cells are unconventional T cells with an important and unique role in antimicrobial immune responses. In healthy adults, MAIT cells are predominantly CD8+ with a smaller DN subset. The phenotypic and functional characteristics of these two MAIT cell subsets, as well as the relationship between them, has not been thoroughly investigated. Here, we report that CD8+ and DN MAIT cells display distinct transcriptional programs and functional characteristics, and we outline their developmental relationship. CD8+ MAIT cells respond more strongly in terms of IFN-γ, TNF, and GrzB production, consistent with their higher basal expression of IL-12R, IL-18R, coactivating receptors, cytotoxic molecules, and the transcription factors Eomes and T-bet. Interestingly, DN MAIT cells produce more IL-17 upon stimulation, consistent with their higher RORγt expression. Moreover, DN MAIT cells display higher propensity for apoptosis, consistent with the enrichment of an apoptosis gene signature. The fetal MAIT cell compartment changes during gestation with expansion of the DN subset and contraction of the CD8+ subset. In addition, the CD8+ MAIT cell pool can give rise to the DN subset in vitro, whereas the opposite process does not occur. Altogether, these findings support a model where DN and CD8+ MAIT cells represent functionally distinct subsets, and the DN MAIT cells are derived from CD8+ MAIT cells (Fig. 7).
Fig. 7.
Properties and relationship of the main MAIT cell subsets.
The higher levels in CD8+ MAIT cells of the activating receptors CD2 and CD9, the transcription factors T-bet and Eomes, as well as the cytotoxic proteins GrzB, Gnly, and Prf, led us to hypothesize that this subset would be superior in their capacity to respond to stimuli. Indeed, peripheral blood CD8+ MAIT cells stimulated with a riboflavin synthesis-competent E. coli strain, or with PMA/ionomycin, produced higher levels of IFN-γ, TNF, and GrzB than their CD8− counterparts. Interestingly, CD8+ MAIT cells maintained their superior functional capacity when stimulated with riboflavin synthesis-incompetent E. coli, and this was likely in part due to higher levels of IL-12R and IL-18R. This distinct responsiveness of CD8+ and DN MAIT cells may have implications in the overall MAIT cell response to microbes, including those that do not synthesize riboflavin. Given the higher responsiveness of peripheral blood CD8+ MAIT cells, it would be beneficial to have highly functional CD8+ MAIT cells strategically located at sites where the likelihood of microbial encounter is higher. Notably, MAIT cells in the rectal mucosa reportedly express higher levels of activation and proinflammatory genes than circulating MAIT cells, with tissue localization influencing their functional repertoire (40), and mucosal CD8+ MAIT cells express higher levels of these genes than their negative counterparts (20). Jo et al. (41) showed that CD8+ MAIT cells were the main producers of IFN-γ within the hepatic MAIT cell compartment in response to TLR8 agonist stimulation.
The basis for higher production of IFN-γ, TNF, and GrzB in CD8+ MAIT cells is unclear. However, it is possible that the CD8 molecule stabilizes and enhances the interaction between the MAIT cell TCR and the MR1–antigen complex, leading to stronger responses, similar to conventional MHC-restricted CD8+ T cells (42, 43). Consistent with this hypothesis, Kurioka et al. (44) showed that blocking CD8 with mAb led to decreased MAIT cell responses against E. coli. CD8-blocking in conventional CD8+ T cells similarly decrease their proliferative capacity, whereas the killing efficiency and IL-2 production are less affected (45). However, our data show that CD8+ MAIT cells are still functionally superior to DN MAIT cells when stimulated with an MR1-independent riboflavin auxotrophic E. coli strain or PMA/ionomycin. Altogether, while CD8 binding to MR1 may influence CD8+ MAIT cell effector functions, other cell-intrinsic or context-dependent mechanisms may also be involved. Of note, higher functional capacity of CD8+ MAIT cells has been previously reported following stimulation of PBMCs with Helicobacter pylori (46) and PMA/ionomycin (47). However, down-regulation of CD8 following stimulation was not considered, suggesting that the DN MAIT cells assessed in those studies may represent a mixture of bona fide DN cells and CD8+ MAIT cells that down-regulated CD8 upon activation.
The mechanism underlying the higher IL-17 production by DN MAIT cells following PMA/ionomycin stimulation is unclear, especially because RORγt expression in CD8+ and DN peripheral blood MAIT cell subsets is similar. This pattern, however, is consistent with higher IL-17 production by endometrial MAIT cells compared with peripheral blood MAIT cells, despite their similar levels of RORγt expression (12). Recent studies reported that IL-7R–, IL-23R–, and STAT3-dependent signaling are important for IL-17 production by MAIT cells (48–50), and thus warrant further investigation in the context of increased IL-17 production by DN MAIT cells. The functional differences between CD8+ and DN MAIT cells are reminiscent of those of subsets of iNKT cells. The circulating iNKT cell population in healthy adults is comprised of CD4+ and DN cells, as well as a minor population of CD8+ cells (51). Although iNKT cells are able to produce both Th1 (IFN-γ and TNF) and Th2 (IL-4, IL-5, and IL-13) cytokines (51, 52), functional differences between the subsets have also been reported, with CD4+ NKT cells being the highest producers of Th2 cytokines (51, 52) and DN NKT cells secreting high levels of Th1 cytokines and IL-17 (52).
MAIT cells express the invariant TCR Vα7.2 segment coupled to the Jα33, Jα12, or Jα20 segments (2–4). However, the Vβ segment usage in total MAIT cells is more diverse (2–4). The present results indicate that DN MAIT cells express a more limited set of Vβ segments than CD8+ MAIT cells. Interestingly, the Vβ repertoire of DN MAIT cells is a limited subrepertoire of the CD8+ MAIT cell Vβ repertoire. Furthermore, analysis of TCR transcripts revealed higher TCR sequence diversity in CD8+ MAIT cells than in DN MAIT cells. These findings, together with the observation that CD8 is down-regulated from the surface upon stimulation, support the notion that DN MAIT cells may represent a subset of CD8+ MAIT cells that responded in vivo, and subsequently down-regulated CD8. We previously showed that fetal CD8αα MAIT cells may be derived from CD8αβ MAIT cells in a stepwise manner in vivo (CD8αβhi → CD8αβlo → CD8αα) (13). Walker et al. (53) previously also suggested that adult peripheral blood CD161++ CD8αα T cells derive from CD161++ CD8αβ T cells. In the present study, further analysis of cells from fetal tissues revealed that fetal DN MAIT cells were more mature than CD8+ MAIT cells, as evaluated by their higher expression of CD45RO and IL-18R, and lower expression of CD62L and CCR7, and expanded during the gestational period, with a corresponding contraction of CD8+ MAIT cells. Moreover, cord blood DN MAIT cells were mature stage 3 CD27+ CD161hi MAIT cells. Interestingly, a recent study showed that DN MAIT cells are rare in the thymus but more abundant in UCB and even more so in adult peripheral blood (33). Altogether, these findings are consistent with the model we propose, whereby the DN MAIT cells appear later during development, potentially following in vivo activation of CD8+ MAIT cells (Fig. 7). Future studies using wild-derived mouse models, such as the CAST/EIJ mice, which have higher MAIT cell frequency than common inbred strains (54), will be critical to further understand the developmental relationship between CD8+ and DN MAIT cells and the precise mechanisms by which DN MAIT cells are generated in vivo.
The DN MAIT cells stained at higher levels with FLICA, a reagent labeling cells undergoing caspase-mediated cell death following bacterial or mitogen stimulation. This indicates that DN MAIT cells are more prone to apoptosis than CD8+ MAIT cells, which is consistent with recent findings (44). This pattern was also in agreement with the enrichment of an apoptosis transcriptional signature, the higher expression levels of Bax in relation to Bcl-2, as well as of PLZF, in the DN MAIT cells (39). Furthermore, when sorted CD8+ MAIT cells were stimulated via their TCR in vitro, the induced DN MAIT cell population developed high expression of activated caspase 3. Because viable DN MAIT cells negative for both FLICA and DCM still had lower functionality than the corresponding CD8+ MAIT cells, the overall lower functionality of DN MAIT cells was likely not caused by lower viability of the DN MAIT cells. Interestingly, a study on chronic hepatitis C virus (HCV) infection showed that the levels of peripheral blood CD8+ MAIT cells were lower in HCV-infected patients compared with healthy controls, but no differences in the levels of DN MAIT cells were detected (24). Similarly, our data indicated that the CD8+ MAIT cells declined during aging and senescence in adult peripheral blood, whereas the DN MAIT cell population remained constant (SI Appendix, Fig. S6F). These observations support a model where DN MAIT cells may be generated from the CD8+ MAIT cell population during steady state or infection at a similar rate at which DN MAIT cells die. Over time, this would result in contraction of the CD8+ MAIT cell population, while the DN MAIT cell compartment remains constant.
In summary, we show marked functional differences between CD8+ and DN MAIT cells, and propose a model where the apoptosis-prone DN MAIT cells may derive from the main CD8+ MAIT cell population (Fig. 7). The realization that CD8+ and DN MAIT cells have distinct functional characteristics will be important for the continued study and understanding of MAIT cells in immune protection of the human host.
Materials and Methods
Human Samples.
Peripheral blood was obtained from healthy individuals at the Blood Transfusion Clinic at the Karolinska University Hospital Huddinge, Sweden. Apheresis samples used for targeted transcriptomic work were collected under protocols (RV229B/WR#1368) with ethical approval provided by the Human Subjects Protection Branch, Walter Reed Army Institute of Research. Uterine tissue samples were collected from women subjected to hysterectomy for nonmalignant and noninflammatory conditions at the St Göran Hospital in Stockholm, Sweden. Diagnosis of human papillomavirus (HPV) infection was performed by the PapilloCheck HPV genotyping test (Greiner Bio-One) at the accredited microbiology laboratory service at the Karolinska University Hospital in Stockholm, Sweden. Umbilical cord blood samples were obtained through the Singapore Cord Blood Bank, from donated units failing to meet the criteria for public clinical banking. Fetal spleens were from second trimester elective abortions from The San Francisco General Hospital Women’s Options Center. Written informed consent was obtained from all donors and ethical approval was obtained from the Regional Ethics Review Board in Stockholm, the Institutional Review Boards of National University of Singapore and Singapore General Hospital, the Research Advisory Ethics Committee of the Singapore Cord Blood Bank, and the Institutional Review Board and the Committee on Human Subjects Research at the University of California, San Francisco.
Blood and Tissue Processing.
PBMCs were isolated by standard Ficoll-Hypaque density gradient separation (Lymphoprep; Axis-Shield). After isolation, PBMCs were cryopreserved until further use, or immediately used for Vα7.2+ and MAIT cell purification or activation assays. Vα7.2+ cells were isolated from PBMCs by positive selection, using anti-Vα7.2 APC-conjugated mAb (Biolegend) and magnetic-activated cell sorting (MACS) anti-APC microbeads, according to the manufacturer’s instructions. Purified Vα7.2+ cells were rested overnight in RPMI-1640 medium supplemented with 25 mM Hepes, 2 mM l-glutamine (all from Thermo Fisher Scientific), 10% FBS (Sigma-Aldrich), 50 μg/mL gentamicin (Life Technologies), and 100 μg/mL normocin (InvivoGen; complete medium). Vα7.2+ cells were subsequently FACS-sorted with CD161 and Vα7.2 mAbs to isolate pure CD8+ and DN MAIT cell populations. In selected experiments, MAIT cells were FACS-sorted directly from PBMCs using the MR1 5-OP-RU tetramers or the CD161 and Vα7.2 mAbs. Monocytes were negatively selected from peripheral blood using the RosetteSep human monocyte enrichment mixture (Stemcell Technologies), as per the manufacturer’s instructions, and rested for 48 h at 37 °C/5% CO2 in complete medium before use in activation assays. UCB mononuclear cells were freshly isolated by density gradient centrifugation (Ficoll-Histopaque Premium; GE Healthcare) as previously described (55). The CD34+ hematopoietic stem and progenitor cells were removed using column-free magnetic beads according to the manufacturer’s instructions (EasySep Human Cord CD34 Positive Selection Mixture; Stemcell Technologies). CD34-depleted UCB mononuclear cells were cryopreserved for further use in 90% (vol/vol) autologous plasma with 10% (vol/vol) DMSO (Sigma Aldrich) (55). The endometrium samples were maintained in ice-cold RPMI-1640 medium supplemented with 50 μg/mL gentamicin and 2.5 μg/mL fungizone (both from Life Technologies), and processed within 24 h after the hysterectomy, as previously described (12). Fetal spleens were collected into cold RPMI medium supplemented with 10% FCS, 25 mM Hepes, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM l-glutamine and maintained on ice. The spleens were processed as previously described (13).
Bacterial Culture.
The E. coli strains 1100-2 and BSV18 were obtained from the Coli Genetic Stock Center, Yale University; the DH5α strain was obtained from New England Biolabs, and the D21 strain was a gift from Peter Bergman, Karolinska Institutet, Stockholm, Sweden. All E. coli strains were grown overnight at 37 °C in Luria (lysogeny) broth (LB), as described previously (56), and supplemented with 20 μg/mL riboflavin (Sigma-Aldrich) for the BSV18 cultures. In selected experiments, bacteria were grown overnight at 37 °C in Riboflavin Assay Medium (RAM; BD Biosciences) in the presence of 3 μg/mL riboflavin dissolved in acetonitrile (Honeywell), or in solvent control alone. Turbidity was measured at 600-nm absorbance using a microplate reader (Infinite M200; Tecan).
MAIT Cell Functional Assays.
MAIT cells within bulk PBMCs were activated using formaldehyde-fixed E. coli strains as indicated, or a combination of IL-12 and IL-18 (10 ng/mL and 100 ng/mL, respectively; PeproTech) for 24 h, as previously described (16, 35). FACS-sorted CD8+ and DN MAIT cells were rested overnight at 37 °C/5% CO2 in complete medium. MAIT cells were then stimulated for 6 h with PMA/ionomycin or for 24 h with formaldehyde-fixed E. coli (strain D21)-fed monocytes (ratio E. coli CFU: monocyte of 30:1 and ratio MAIT cell: monocyte of 1:1) and in the presence of 1.25 μg/mL anti-CD28 mAb (L293; BD Biosciences), and 20 μg/mL MR1 blocking mAb (26.5; Biolegend) or IgG2a isotype control (ctrl) (MOPC-173; Biolegend), as described previously (35). E. coli was mildly fixed in formaldehyde as previously described (56). In all activation assays, monensin (Golgi Stop; BD Biosciences) and brefeldin (Golgi Plug; BD Biosciences) were added for the last 6 h of incubation. In selected experiments, MAIT cells were stimulated with PMA/ionomycin for 24 h in the absence of protein transport inhibitors. Cytokines in the supernatants were quantified using the LEGENDplex kit (Biolegend), as per the manufacturer’s instructions. Apoptosis in activated MAIT cells was assessed by using the FLICA reagent (Vybrant FAM Poly Caspases Assay Kit; Thermo Fisher Scientific) as previously described (35), or by staining using antiactive caspase 3 mAb (SI Appendix, Table S4). In the derivative experiments, FACS-sorted CD8+ and DN MAIT cells prestained with 1.25 μM CTV (Thermo Fisher Scientific) were incubated for up to 7 d in culture plates that had been precoated with 10 μg/mL Vα7.2 mAb (3C10; Biolegend) and CD28 mAb (CD28.2; Biolegend).
Flow Cytometry.
Cell surface and intracellular staining for cytokines, cytotoxic molecules, transcription factors, and staining using the LEGENDScreen kit (Biolegend) were performed as previously described (56). Staining with the MR1 5-OP-RU and MR1 6-FP tetramers conjugated with PE were performed for 40 min at room temperature (10) before proceeding to the surface staining with other mAbs (SI Appendix, Table S4). Samples were acquired on an LSRFortessa flow cytometer (BD Biosciences) equipped with 355-, 405-, 488-, 561, and 639-nm lasers. FACS-sorting was performed on FACSAriaIII or FACSAria Fusion equipped with 355-, 405-, 488-, 561-, and 640-nm lasers, or FACSMelody equipped with 405-, 488-, and 640-nm lasers (all BD Biosciences). Single-stained polystyrene beads (BD Biosciences) and the compensation platform in FACSDiva v. 8.0.1 (BD Biosciences) or FlowJo software v. 9.9 (TreeStar) were used for compensation.
Gene-Expression Analysis.
CD8+ and DN MAIT cells were FACS-sorted at 100 cells per well into 96-well plates containing a reverse-transcription preamplification reaction mix, which contains 0.2× assay mix [85 preselected primers and probes (SI Appendix, Table S2); Thermo Fisher Scientific], SuperScript III platinum Taq (Thermo Fisher Scientific), and SUPERase-In RNase Inhibitor (Thermo Fisher Scientific). The reverse-transcription reaction was performed as 1 cycle of 15 min at 50 °C followed by 2 min at 95 °C, and preamplification was 16 cycles of 95 °C for 15 s, and 60 °C for 4 min. cDNA produced following this step was diluted 1:5 in DNA suspension buffer (Teknova). Diluted cDNA was combined with TaqMan Universal PCR Master mix (Thermo Fisher Scientific) and further diluted with 20× GE sample loading reagent (Fluidigm), and loaded into one side of a primed, 96.96 Dynamic Array chip (Fluidigm). Experimental assays (20× assay mixes of TaqMan primers and probes) were diluted 1:1 with 2× assay loading buffer (Fluidigm), and loaded onto the other side of the chip. The chip was loaded into an IFC Controller (Fluidigm) to fill the chip matrix with sample and assay, and then transferred to Biomark (Fluidigm) for thermocycling and fluorescence acquisition using the GE 96 × 96 standard v1 program. For quality control, qPCR amplification curves were first validated using gene expression Fluidigm Biomark real-time PCR analysis software (Fluidigm).
RNA Sequencing.
CD8+ and DN MAIT cells from peripheral blood (2000 cells per subset) were sorted using a FACSAriaII (BD Biosciences) directly into lysis buffer provided with the SMART-Seq v4 Ultra Low Input RNA Kit (Takara), and then snap frozen. RNAseq libraries were prepared using the SMART-Seq v4 Ultra Low Input RNA Kit according to the manufacturer’s instructions. Briefly, 3′ SMART-Seq CDS (oligo-dt) primers were hybridized to the poly(A) tail of all of the mRNA and reverse-transcribed with the SMARTScribe RT enzyme. cDNA was preamplified using SeqAmp DNA polymerase and frozen. Amplified material was purified using Agencourt AMPure XP beads (Beckman). cDNA quantity was assessed on a Qubit 3.0 (Thermo) and fragment size evaluated on a 2100 BioAnalyzer (Agilent). The PCR products were next indexed using the Nextera XT DNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. Briefly, products were tagmented using the Amplicon tagment mix containing Tn5 transposase, and indexed using Nextera index 1 (i7) and index 2 (i5) primers. The libraries were again cleaned-up with Agencourt AMPure XP beads, pooled, quantified, and sequenced across 75 bp using a paired-end strategy with a 150-cycle high-output flow cell on a NextSeq 550 (Illumina). Three biological replicates were sequenced per experiment.
RNAseq Analysis.
Fastq files from three sequencing runs were concatenated and aligned using STAR 2.5.2a and hg38 to generate a final unique paired mapped read depth ranging from 8 million to 13.6 million reads per sample. The aligned files were normalized using Deseq2. Differentially expressed genes on normalized RNAseq counts between bulk CD8+ and DN MAIT cells were assessed using a t test (P < 0.05) with the R limma package (v3.28.21). Normalized RNAseq counts from sorted CD8+ and DN MAIT cells were subjected to GSEA using Broad Institute software (software.broadinstitute.org/gsea/index.jsp). The TCR data were extracted from the RNAseq dataset using the MiXCR software (https://mixcr.readthedocs.io/en/master/index.html).
Data Availability.
The sequence datasets have been deposited in the Gene Expression Omnibus with the accession no. GSE120847.
Statistical Analyses.
In gene-expression analyses, volcano plots representing gene expression fold-change and P value (Wilcoxon signed rank test) were performed in R. The P values were corrected for multiple comparisons using the Benjamini and Hochberg (57) false-discovery rate (FDR) method and selected genes were filtered based on a FDR < 0.1. The remaining statistical analyses were performed using Prism software v6 (GraphPad). The paired t test or Wilcoxon’s signed-rank test, and the unpaired t test were used as appropriate to detect significant differences between paired and unpaired samples, respectively. The Friedman test followed by Dunn’s multiple comparison test was used to determine significance across multiple, paired samples. The Spearman’s rank correlation was used for correlation analyses. Two-sided P < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Åsa-Lena Dackland and Dr. Iyadh Douagi (Karolinska Institutet) and Charles K. L. Lou and Charlene S. F. Foong (Singapore Health Services) for their technical assistance with FACS-sorting experiments; and Dohoon Kim (Walter Reed Army Institute of Research) for assisting in Fluidigm Biomark gene-expression analyses. The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie; and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This research was supported by Swedish Research Council Grant 2015-00174; Marie Skłodowska Curie Actions; Cofund; Project INCA 600398; the Jonas Söderquist Foundation for Virology and Immunology; the Erik and Edith Fernström Foundation for Medical Research; the Swedish Society of Medicine (E.L.); and Swedish Research Council Grant 2016-03052, Swedish Cancer Society Grant CAN 2017/777, and National Institutes of Health Grant R01DK108350 (to J.K.S.). J.D. was supported by Fundação para a Ciência e a Tecnologia Doctoral Fellowship SFRH/BD/85290/2012, cofunded by the Programa Operacional Potencial Humano-Quadro de Referência Estratégico Nacional and the European Social Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE120847).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812273115/-/DCSupplemental.
References
- 1.Fergusson JR, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014;9:1075–1088. doi: 10.1016/j.celrep.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lepore M, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun. 2014;5:3866, and correction (2014) 5:4493. doi: 10.1038/ncomms4866. [DOI] [PubMed] [Google Scholar]
- 3.Reantragoon R, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilloy F, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 6.Dusseaux M, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 7.Martin E, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci USA. 2017;114:E5434–E5443. doi: 10.1073/pnas.1705759114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–708, and correction (2010) 11:969. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 10.Corbett AJ, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–365. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- 11.Kjer-Nielsen L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 12.Gibbs A, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017;10:35–45. doi: 10.1038/mi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurioka A, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 2015;8:429–440. doi: 10.1038/mi.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Bourhis L, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 2013;9:e1003681. doi: 10.1371/journal.ppat.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leeansyah E, et al. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. 2015;11:e1005072. doi: 10.1371/journal.ppat.1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salou M, Franciszkiewicz K, Lantz O. MAIT cells in infectious diseases. Curr Opin Immunol. 2017;48:7–14. doi: 10.1016/j.coi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Loh L, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci USA. 2016;113:10133–10138. doi: 10.1073/pnas.1610750113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ussher JE, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2014;44:195–203. doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slichter CK, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. 2016;1:e86292. doi: 10.1172/jci.insight.86292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ussher JE, et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur J Immunol. 2016;46:1600–1614. doi: 10.1002/eji.201545969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beudeker BJB, et al. Mucosal‐associated invariant T-cell frequency and function in blood and liver of HCV mono- and HCV/HIV co-infected patients with advanced fibrosis. Liver Int. 2017;38:458–468. doi: 10.1111/liv.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeijen LLM, et al. Mucosal-associated invariant T cells are more activated in chronic hepatitis B, but not depleted in blood: Reversal by antiviral therapy. J Infect Dis. 2017;216:969–976. doi: 10.1093/infdis/jix425. [DOI] [PubMed] [Google Scholar]
- 24.Bolte FJ, et al. Intra-hepatic depletion of mucosal-associated invariant T cells in hepatitis C virus-induced liver inflammation. Gastroenterology. 2017;153:1392–1403.e2. doi: 10.1053/j.gastro.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove C, et al. Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood. 2013;121:951–961. doi: 10.1182/blood-2012-06-436436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengst J, et al. Nonreversible MAIT cell-dysfunction in chronic hepatitis C virus infection despite successful interferon-free therapy. Eur J Immunol. 2016;46:2204–2210. doi: 10.1002/eji.201646447. [DOI] [PubMed] [Google Scholar]
- 27.Leeansyah E, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121:1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wilgenburg B, et al. STOP-HCV consortium MAIT cells are activated during human viral infections. Nat Commun. 2016;7:11653. doi: 10.1038/ncomms11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandberg JK, Norrby-Teglund A, Leeansyah E. Bacterial deception of MAIT cells in a cloud of superantigen and cytokines. PLoS Biol. 2017;15:e2003167. doi: 10.1371/journal.pbio.2003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaler CR, et al. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: Defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. 2017;15:e2001930. doi: 10.1371/journal.pbio.2001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gold MC, et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 2013;6:35–44. doi: 10.1038/mi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seach N, et al. Double-positive thymocytes select mucosal-associated invariant T cells. J Immunol. 2013;191:6002–6009. doi: 10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- 33.Koay HF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17:1300–1311. doi: 10.1038/ni.3565. [DOI] [PubMed] [Google Scholar]
- 34.Gherardin NA, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. 2018;96:507–525. doi: 10.1111/imcb.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dias J, Sobkowiak MJ, Sandberg JK, Leeansyah E. Human MAIT-cell responses to Escherichia coli: Activation, cytokine production, proliferation, and cytotoxicity. J Leukoc Biol. 2016;100:233–240. doi: 10.1189/jlb.4TA0815-391RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bandrin SV, Rabinovich PM, Stepanov AI. [3 linkage groups of the genes of riboflavin biosynthesis in Escherichia coli] Genetika. 1983;19:1419–1425. Russian. [PubMed] [Google Scholar]
- 37.Bennett MS, Trivedi S, Iyer AS, Hale JS, Leung DT. Human mucosal-associated invariant T (MAIT) cells possess capacity for B cell help. J Leukoc Biol. 2017;102:1261–1269. doi: 10.1189/jlb.4A0317-116R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 39.Gérart S, et al. Human iNKT and MAIT cells exhibit a PLZF-dependent proapoptotic propensity that is counterbalanced by XIAP. Blood. 2013;121:614–623. doi: 10.1182/blood-2012-09-456095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dias J, et al. Factors influencing functional heterogeneity in human mucosa-associated invariant T cells. Front Immunol. 2018;9:1602. doi: 10.3389/fimmu.2018.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jo J, et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. 2014;10:e1004210. doi: 10.1371/journal.ppat.1004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norment AM, Salter RD, Parham P, Engelhard VH, Littman DR. Cell-cell adhesion mediated by CD8 and MHC class I molecules. Nature. 1988;336:79–81. doi: 10.1038/336079a0. [DOI] [PubMed] [Google Scholar]
- 43.Salter RD, et al. A binding site for the T-cell co-receptor CD8 on the alpha 3 domain of HLA-A2. Nature. 1990;345:41–46. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- 44.Kurioka A, et al. Shared and distinct phenotypes and functions of human CD161++ Vα7.2+ T cell subsets. Front Immunol. 2017;8:1031. doi: 10.3389/fimmu.2017.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caveno J, Zhang Y, Motyka B, Teh SJ, Teh HS. Functional similarity and differences between selection-independent CD4-CD8- alphabeta T cells and positively selected CD8 T cells expressing the same TCR and the induction of anergy in CD4-CD8- alphabeta T cells in antigen-expressing mice. J Immunol. 1999;163:1222–1229. [PubMed] [Google Scholar]
- 46.Booth JS, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood: Role in Helicobacter pylori infection. Front Immunol. 2015;6:466. doi: 10.3389/fimmu.2015.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brozova J, Karlova I, Novak J. Analysis of the phenotype and function of the subpopulations of mucosal-associated invariant T Cells. Scand J Immunol. 2016;84:245–251. doi: 10.1111/sji.12467. [DOI] [PubMed] [Google Scholar]
- 48.Guggino G, et al. IL-17 polarization of MAIT cells is derived from the activation of two different pathways. Eur J Immunol. 2017;47:2002–2003. doi: 10.1002/eji.201747140. [DOI] [PubMed] [Google Scholar]
- 49.Willing A, Jäger J, Reinhardt S, Kursawe N, Friese MA. Production of IL-17 by MAIT cells is increased in multiple sclerosis and is associated with IL-7 receptor expression. J Immunol. 2018;200:974–982. doi: 10.4049/jimmunol.1701213. [DOI] [PubMed] [Google Scholar]
- 50.Wilson RP, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. 2015;212:855–864. doi: 10.1084/jem.20141992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan AC, et al. Ex-vivo analysis of human natural killer T cells demonstrates heterogeneity between tissues and within established CD4(+) and CD4(-) subsets. Clin Exp Immunol. 2013;172:129–137. doi: 10.1111/cei.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Reilly V, et al. Distinct and overlapping effector functions of expanded human CD4+, CD8α+ and CD4-CD8α- invariant natural killer T cells. PLoS One. 2011;6:e28648. doi: 10.1371/journal.pone.0028648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker LJ, et al. Human MAIT and CD8αα cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119:422–433. doi: 10.1182/blood-2011-05-353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui Y, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. 2015;125:4171–4185. doi: 10.1172/JCI82424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu PP, et al. Intercellular cytosolic transfer correlates with mesenchymal stromal cell rescue of umbilical cord blood cell viability during ex vivo expansion. Cytotherapy. 2012;14:1064–1079. doi: 10.3109/14653249.2012.697146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dias J, Sandberg JK, Leeansyah E. Extensive phenotypic analysis, transcription factor profiling, and effector cytokine production of human MAIT cells by flow cytometry. Methods Mol Biol. 2017;1514:241–256. doi: 10.1007/978-1-4939-6548-9_17. [DOI] [PubMed] [Google Scholar]
- 57.Benjamini Y, Hochberg C. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence datasets have been deposited in the Gene Expression Omnibus with the accession no. GSE120847.