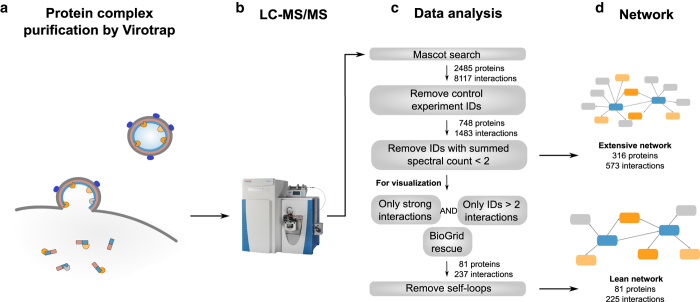
Figure 2. Workflow to generate the protein-protein interaction dataset.
(a) Gag-bait fusion proteins were expressed in HEK293T cells and virus-like particles, trapping the protein complexes, were purified and prepared for subsequent LC-MS/MS analysis (b). Three biological replicates for every bait protein were performed, both for experiments without and with TNF treatment. (c) The MS/MS-data were searched with MASCOT and resulted in 2485 identified proteins. All proteins identified in a control experiment were removed, resulting in 748 remaining proteins. Interacting proteins identified with a single PSM were removed, resulting in an extensive network (d) with 316 proteins and 573 interactions. For network visualization, the dataset was further filtered (c). Only strong interactions (based on spectral counts) and interacting proteins with more than three interactions were withheld, unless the interacting protein was a known interactor in BioGRID. (d) The lean network consists of 81 proteins and 225 interactions.