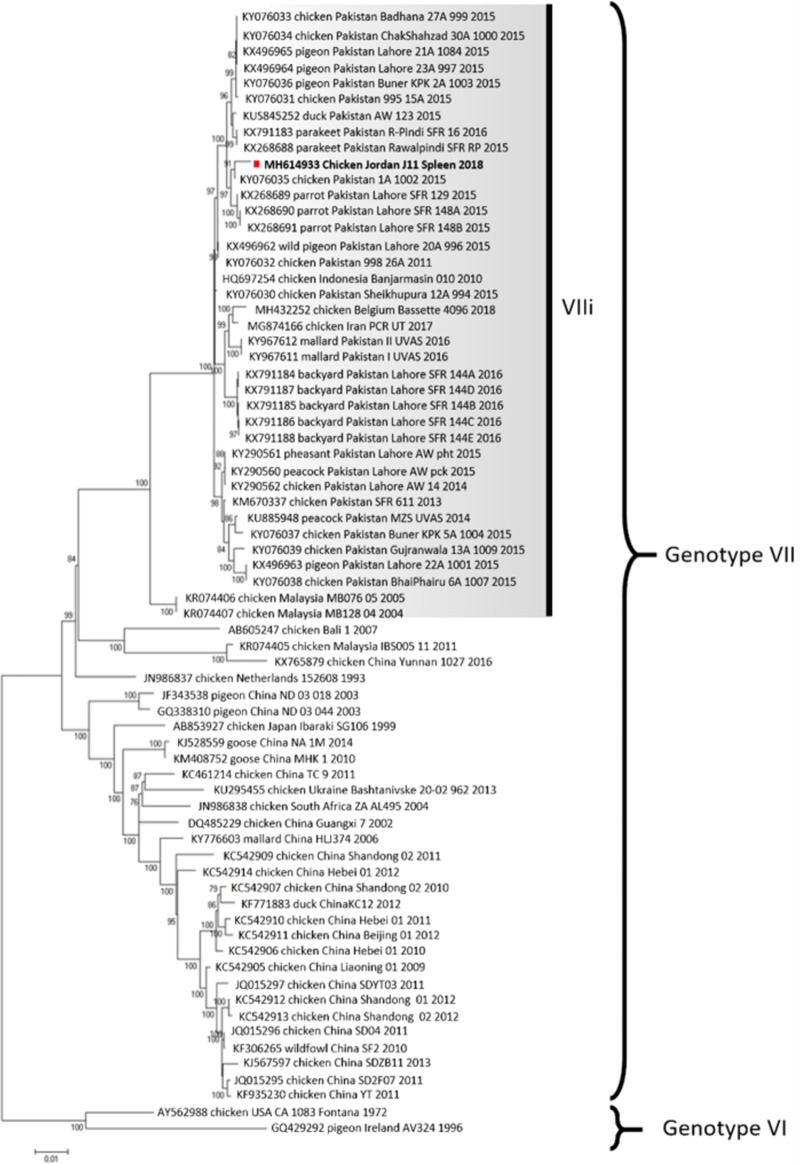
FIG 1.
Evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model. The tree with the highest log likelihood (−61,001.55) is shown. Percentage of trees in which the associated taxa clustered together is shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying Neighbor-Join (NJ) and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with the superior log-likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.3664]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Analysis involved 70 nucleotide sequences. Codon positions included were first, second, third, and noncoding. All positions containing gaps and missing data were eliminated. A total of 13,744 positions were in the final data set of concatenated coding sequences of 6 NDV genes. The sample NDV/chicken/Jordan/J11-spleen/2018 sequence is highlighted with a red square.