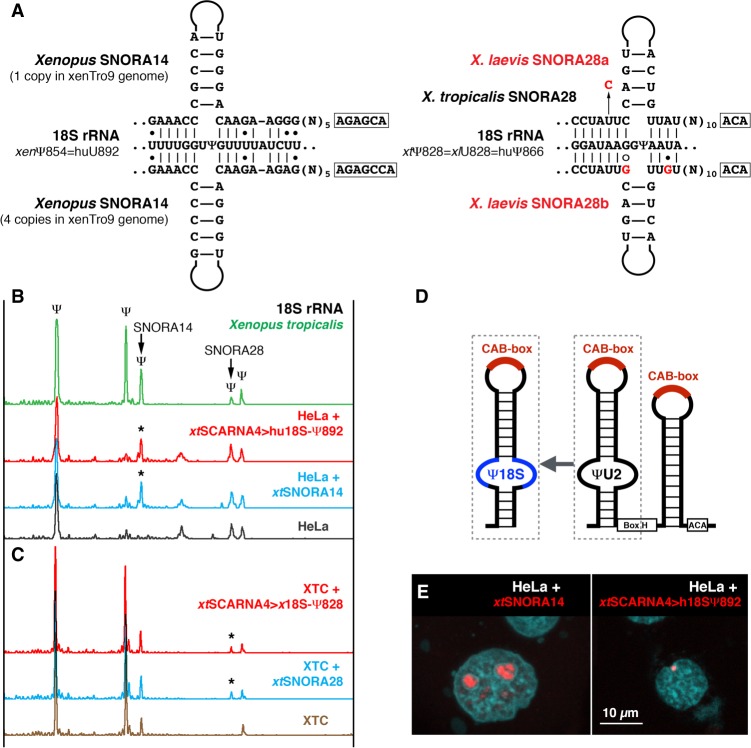
FIGURE 3.
Typical scaRNAs can modify 18S rRNA. (A) Postulated base-pairing of Xenopus snoRNAs, SNORA14, and SNORA28, with 18S rRNA. Differences between X. tropicalis and X. laevis SNORA28 are highlighted in red. (B) 18S rRNA from X. tropicalis is pseudouridylated by SNORA28 at position 828 and by SNORA14 at position 854 (top green trace). In HeLa cells, the position corresponding to the Xenopus SNORA14 target is not modified (bottom gray trace). When Xenopus SNORA14 was expressed in HeLa cells, position 892 (equivalent to position 854 in Xenopus) became pseudouridylated (blue trace, star). Human 18S-892 guide RNA made from Xenopus SCARNA4 (xtSCARNA4>hu18S-Ψ892) also induced the pseudouridylation of 18S rRNA in HeLa cells (red trace, star). (C) In contrast to human and X. tropicalis, 18S rRNA in X. laevis is not pseudouridylated at position 828 (bottom brown trace). Expression of X. tropicalis SNORA28 in XTC cells induced pseudouridylation of this position (blue trace, star). This pseudouridine was also induced by the chimeric scaRNA xtSCARNA4>x18S-Ψ828 (red trace, star). (D) Schematic representation of 18S guide RNA generated from scaRNA. (E) In transfected HeLa cells, FISH with Cy3-labeled antisense RNA probes shows that xtSNORA14 localizes in nucleoli (left panel, red signal) and xtSCARNA4>hu18SΨ892 localizes in CBs (right panel, red signal).