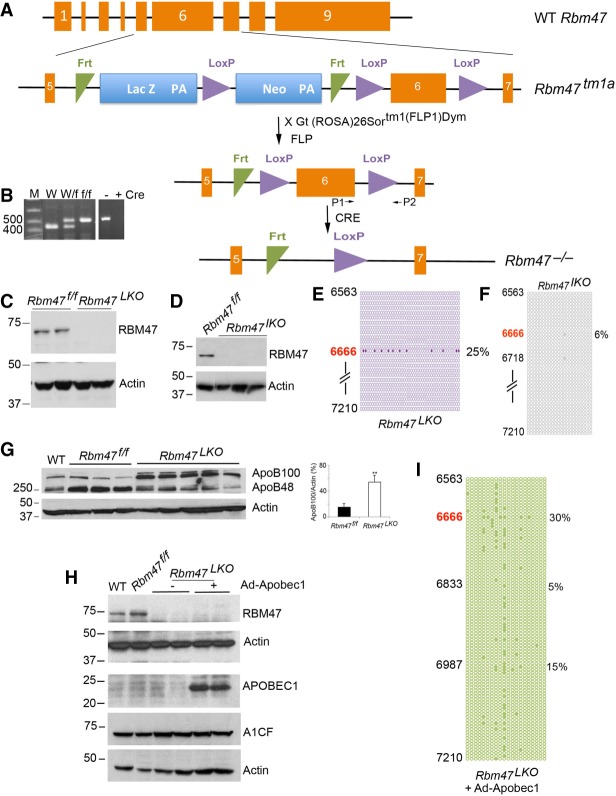
FIGURE 3.
Generation and characterization of conditional Rbm47 knockout mice. (A) Endogenous mouse Rbm47 gene and targeting construct. Schematic illustration of genomic structure of Rbm47 allele containing nine exons (orange boxes). Schematic of Rbm47tm1a composition. (Numbered oranges boxes) Rbm47 exons; (blue boxes) Lac Z and neo cassettes; (green triangles) FRT sites; (purple triangles) LoxP sites. FLP recombination eliminates the LacZ and neo cassettes. Exon 6 containing transcriptional start codon and sequences encoding RNA recognition motifs was targeted for Cre-dependent homologous recombination. (B) Representative DNA electrophoresis showing genotype of WT and targeted alleles using primers P1 and P2 surrounding the 3′arm LoxP site. Analysis of the WT allele results in a 490 bp PCR product (lane 1) whereas recombination at the floxed allele generates a 530 bp product (f/f) (lane 3). Cre activation resuts in the loss of exon 6, evidenced by the absence of PCR amplification (lane 5). Molecular weights (bp) are indicated to the left. (C) Western blot detection of RBM47 in liver of floxed and Rbm47 liver-specific knockout (Rbm47LKO) mice with Actin used as loading control. (D) Western blot analysis of RBM47 in Rbm47IKO mice showing no detectable protein in intestinal nuclear extract. (E) Hepatic apoB RNA editing profile in Rbm47LKO mice, representative of three individual livers. Forty-two clones were sequenced, with each clone represented by a circle. Solid circles indicate editing at the specified position. We examined an apoB region encompassing nucleotides 6563–7210, with editing only at the canonical cytidine (6666) detected. (F) Intestinal apoB RNA editing profile in Rbm47IKO mice. Representative distribution of RNA editing sites from the same region (6563–7210) revealing only 2/19 clones exhibiting RNA editing. (G) Western blot analysis of hepatic apoB100 and apoB48 isoforms (three to five mice per genotype) with actin as loading control. (Panel to the right ) Quantitation of apoB100 isoform as a fraction of total apoB showing significant increase of apoB100 in liver of Rbm47LKO mice. (H) Western blot analysis of RBM47 and A1CF in Rbm47LKO mice at baseline and following adenoviral APOBEC1 overexpression. (I) Editing and hyperediting profile of hepatic apoB RNA following adenoviral overexpression of APOBEC1 in two Rbm47LKO mice. Representative distribution of edited sites from nucleotide 6563 to nucleotide 7210.