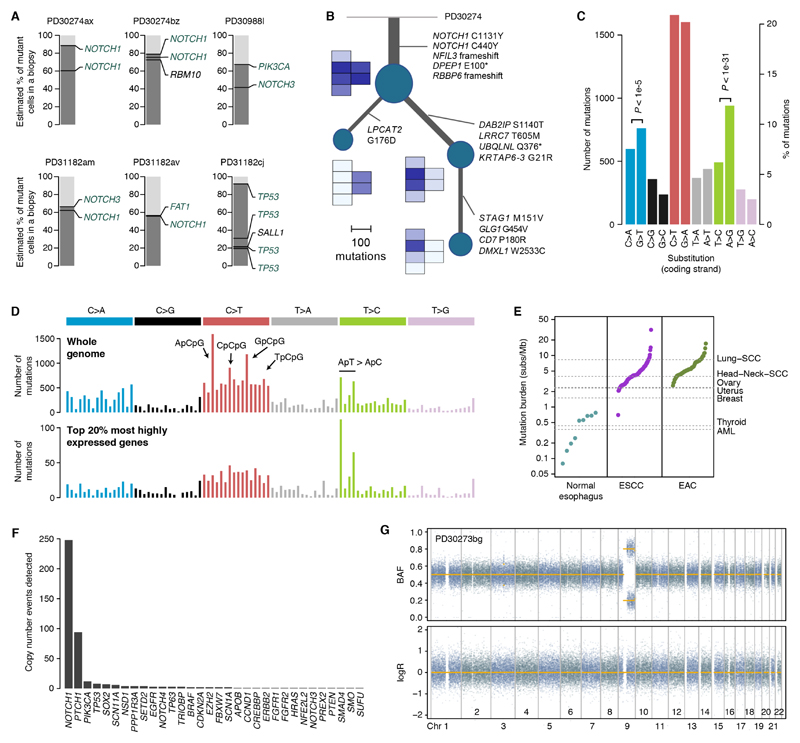
Fig. 4. Phylogenetic and mutational patterns in normal esophagus.
(A) Representation of mutations co-occurring in the same clones using the pigeonhole principle (see Supplementary material 5.5). (B) Phylogenetic reconstruction of the evolution of a large clone overlapping six samples using whole-genome sequencing data and spatial information. A small heatmap of the six affected samples is shown next to each node in the tree, depicting the mean VAF of the mutations in each node. (C) Number of substitutions per mutation type as mapped to the coding (untranscribed) strand from all donors. P-values reflect transcription strand asymmetry (exact Poisson test). (D) 96-mutation-class barplot depicting the number of mutations in each of the possible 96 trinucleotides (strand independent). The top panel shows the whole-genome plot aggregating all 21 whole-genomes. The bottom panel shows the spectrum for mutations occurring in the transcribed region of the top 20% most highly expressed genes. (E) Mutation burden in normal esophagus and in ESCC and EAC tumors (every point corresponds to a donor, sorted by mutation burden). (F) Number of copy number events detected in each gene across the 844 samples using the targeted data. (G) Representative LogR and B-allele frequency (BAF) scatter plots for heterozygous SNPs from whole genome data showing a copy-neutral LOH event affecting NOTCH1 (sample PD30273bg shown).