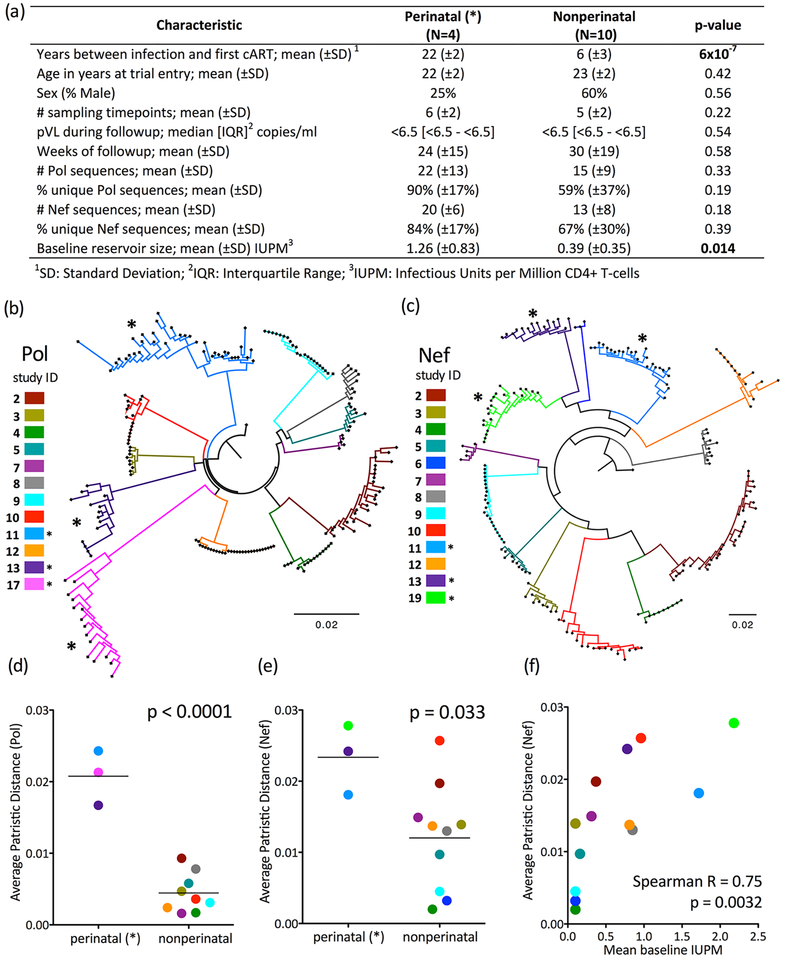
Figure 1: Genetic diversity within the replication-competent latent HIV reservoir increases with untreated infection duration.
Panel A: Participant clinical, immunogenetic, and HIV reservoir dataset characteristics. Throughout all figures, perinatally-infected participants (those with longer uncontrolled infection duration) are denoted by asterisks (*). Panel B: Maximum-likelihood phylogeny relating within-host HIV Pol sequences, colored by participant. All within-host sequence datasets formed monophyletic clades with 100% bootstrap support. Scale in expected nucleotide substitutions per site. Panel C: same as B, but for Nef. All within-host datasets formed monophyletic clades with 100% bootstrap support except those of participants 11 (99%) and 19 (89%). Differences in overall topologies between Pol and Nef trees are attributable to low bootstrap support for the deeper branches (i.e. those that define evolutionary relationships between participants); all were <70% except the subclade comprising participants 2 and 4. No downstream analyses however relied on intra-participant genetic distances. Panel D: Average within-host patristic (tip-to-tip phylogenetic) distances in reservoir Pol sequences; p-value calculated using Student’s T-test. Panel E: Same as panel D, but for Nef. Panel F: Relationship between size and diversity (Nef) of the within-host replication-competent HIV reservoir, assessed by Spearman’s correlation.