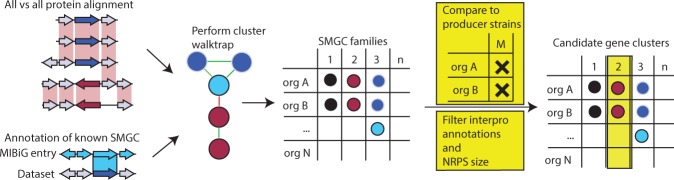
Figure 1.
Workflow of the bioinformatic pipeline. Prior to data analysis gene annotation, InterPro and SMURF data are combined. SMGC are compared using protein BLAST of cluster members and percent identity values of alignments are aggregated to cluster similarity scores and used to create a gene cluster network. Additionally, known gene clusters from the MIBiG database are annotated in the dataset by identifying an exact match. Random walk clustering is performed using the cluster walktrap function52 of igraph51 on the network to obtain families of SMGC. To identify candidate SMGC for metabolites of interest, lists of metabolite producing organisms are compared to lists of organisms containing SMGCs of the same family. Candidate SMGC families are filtered by interpro annotations and e.g. NRPS size.