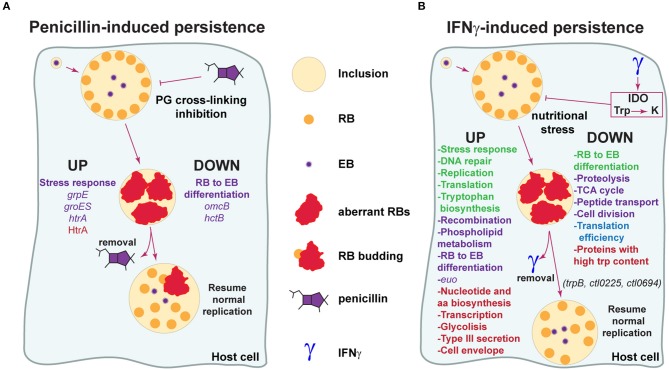
Figure 2.
Model for the molecular basis of Chlamydia evasion of antimicrobial activity elicited by penicillins and IFNγ. (A) Penicillins inhibit crosslinking of the peptidoglycan (PG), leading to disruption of cell wall synthesis. Chlamydia respond to this stress by downregulating genes required for RB to EB transition (omcB, hctB), halting cell division and adopting an aberrant, enlarged morphology (aberrant RB) (Goellner et al., 2006; Ouellette et al., 2006). Genes associated to stress responses are upregulated (grpE, groES, htrA) (Goellner et al., 2006; Di Pietro et al., 2012). During this stage, the bacteria is able to “persist” for long periods of time in a viable but non-cultivable state and infectious progeny generation is abrogated. When penicillin is removed, Chlamydia exit persistence and resume generation of normal RBs, which may originate from aRBs through a budding-like process. The protease/chaperone protein HtrA may participate in reversion from penicillin-induced persistence (Huston et al., 2008; Ong et al., 2013). Reported processes/genes/proteins up- and down-regulated during penicillin-induced persistence are indicated. Data based on transcriptional or protein studies are highlighted in purple and red, respectively. (B) IFNγ is a cytokine with key roles in host defense against pathogens, including Chlamydia. IFNγ activates indoleamine-2,3-dioxygenase (IDO), causing the degradation of tryptophan (Trp) into kynurenine (K) and triggering nutritional stress in Chlamydia. In response to this stress, Chlamydia enters into a persistent state analogous to what is observed with penicillin. In this state, Chlamydia alters gene transcription and expression in order to modify key biological processes and warrant survival. Reported processes/genes/proteins up- and down-regulated during IFNγ-induced persistence are indicated. Data based on transcriptional studies (Byrne et al., 2001; Belland et al., 2003; Goellner et al., 2006; Ouellette et al., 2006) is indicated in purple. Data based on protein studies (Molestina et al., 2002; Mukhopadhyay et al., 2006; Ouellette et al., 2006; Østergaard et al., 2016) is shown in red. Data supported by both transcriptional and protein studies is indicated in green. Data based on global activity of the translation pathway is shown in blue. Note that RB to EB differentiation transcripts have been found increased and decreased in different studies, as discussed in the manuscript. trpB (tryptophan synthase), ctl0225 (a predicted small neutral amino acid transporter) and ctl0694 (a hypothetical oxidoreductase) have been found to play a role in reversion from IFNγ-induced persistence in C. trachomatis (Muramatsu et al., 2016).