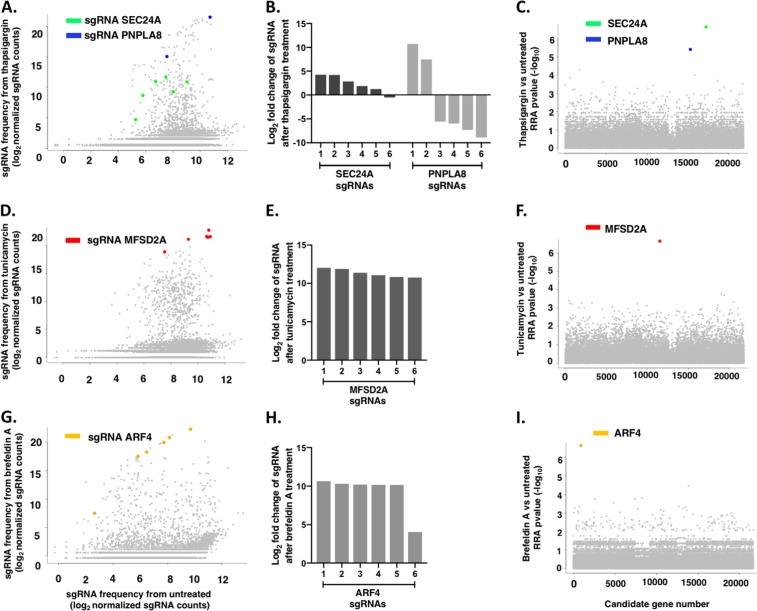
Fig. 1. Genes isolated from positive selection screens in CRISPR/Cas9- modified HAP1 cells using ER stressors.
Each gene is targeted by a total of six sgRNAs; therefore, a screen showing a robust survival phenotype will show multiple sgRNAs targeting the same gene. Scatterplots showing the enrichment of sgRNAs in cells treated with a thapsigargin (0.062 µg/mL), d tunicamycin (0.2 µg/mL), and g brefeldin A (0.045 µg/mL), compared to the representation of these sgRNAs in untreated cells. b, e, h Bar graphs showing fold changes of sgRNAs targeting candidate genes after compound treatment relative to untreated cells. c, f, i Identification of top candidate genes in each screen using RRA p value from MAGeCK analyses