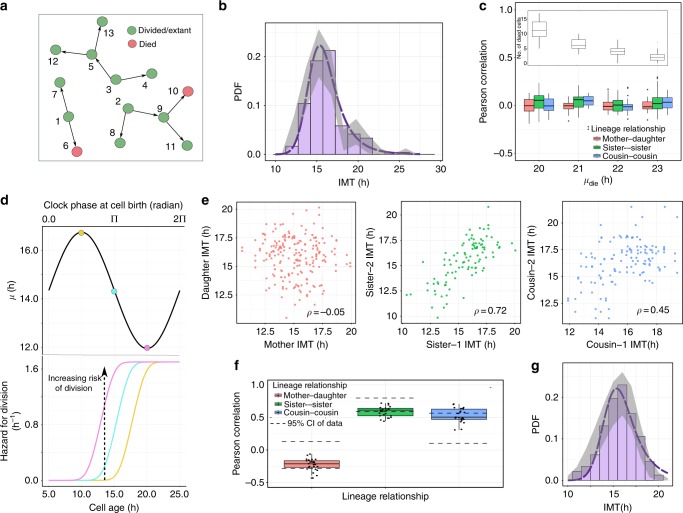
Fig. 5.
Coupling of the cell cycle to the circadian rhythm is required to explain correlations in the absence of cisplatin. a Birth-death process simulations keeping track of lineage relationships. Three ancestor cells and their progeny are shown here as examples. Directed edges represent mother–daughter relationships. b, c Results of a null model with no circadian gating. b The IMT distribution before cisplatin (purple dashed line, EMG parameters in Supplementary Table 4) is almost identical to the experimental data (histogram, EMG parameters in Supplementary section 4). The gray shaded area represents 95% confidence intervals generated from 1000 bootstrap samples of the data. c Pearson correlations between cell pairs as μdie of the EMG function is varied. The inset shows the number of dead cells in 25 simulation runs, for different values of μdie. d–g Model incorporating circadian gating of the cell cycle and results. d A model for the coupling of the circadian clock to the cell cycle. As the phase of the clock at cell birth varies between 0 and 2π, the parameter μ of the EMG function for division varies, thereby controlling the hazard for cell division (top). Three hazard functions corresponding to three phases of the clock are shown in matched colors (bottom), modeling different division risks for three cells born at different phases of the clock. e Instances of Pearson correlations in IMT (ρ) generated from the model with circadian gating of the cell cycle. f The cousin–mother inequality and the magnitude of all lineage correlations are recapitulated with the model for circadian gating of the cell cycle. The dashed lines indicate the 95% confidence intervals of the IMT correlations as measured from the data. g The histogram represents one example of the IMT distribution generated by our simulations incorporating circadian gating. Gray shaded area represents 95% confidence intervals generated from 500 simulation runs. The dashed line represents the inferred IMT distribution, as in b. All parameters used for simulated results in e–g are provided in Supplementary section 6. All boxplots represent the 1st, 2nd, and 3rd quartiles of the lineage correlations generated from 25 simulation runs