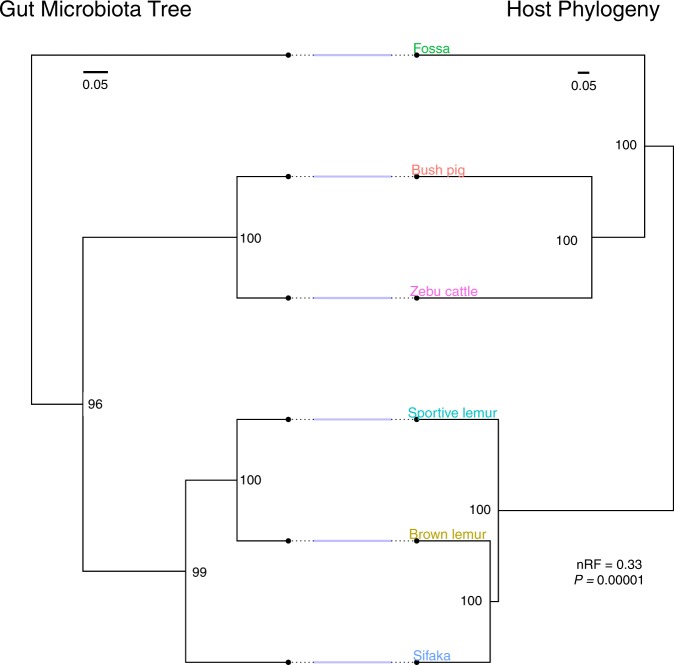
Fig. 3.
Comparison between gut microbiota tree and host phylogeny relationships. The gut microbiota tree reflects the same branching order as the host phylogeny for distantly related host species, but gut microbial similarity among sportive lemurs, brown lemurs, and sifaka does not match these hosts’ established evolutionary relationships. The maximum parsimony gut microbiota tree (left) is based on the abundances of 16S amplicon sequence variants (ASVs) in host species-aggregated microbiome samples. The maximum likelihood host species phylogeny (right) is based on mitochondrial cytochrome b (cyt-b) gene sequences. Topological congruence was quantified using the normalized Robinson-Foulds (nRF) metric, which calculates symmetry in rooted tree shape on a scale from 0.0 (complete congruence) to 1.0 (complete incongruence). The host phylogeny scale bar indicates nucleotide substitutions per site