Fig. 1.
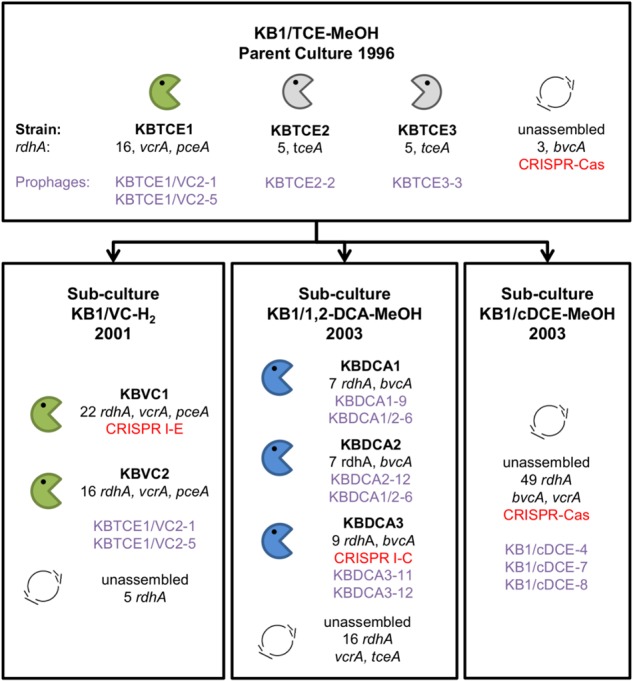
Overview of the eight Dehalococcoides mccartyi genomes closed from the metagenomes of KB-1 enrichment cultures. Enrichment sub-cultures are named by electron acceptor, electron donor, and the date the sub-culture was first created. Electron acceptors include trichloroethene (TCE), vinyl chloride (VC), 1,2-dichloroethane (1,2-DCA), and cis-dichloroethene (cDCE) and donors are either methanol (MeOH) or hydrogen. Closed D. mccartyi genomes were assigned a strain name based on electron acceptor and rank abundance in the mixed culture. The number of reductive dehalogenase homologous genes (rdhA) per strain genome is indicated and any rdhA that has been functionally characterized is listed by name (vcrA, pceA, tceA, and bvcA). Prophages identified in each genome are listed by name in purple. CRISPR-Cas systems are also indicated in red
