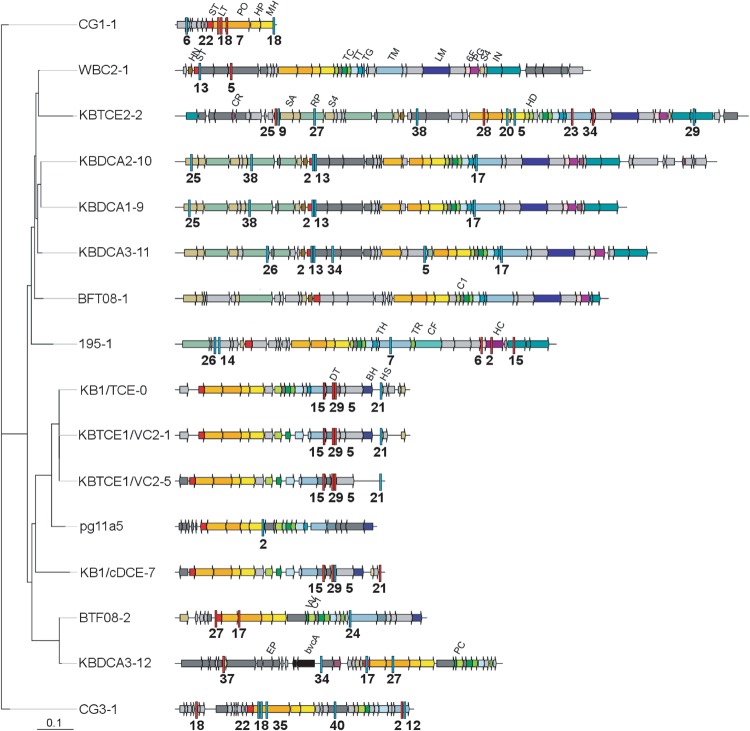
Fig. 2.
Maximum likelihood phylogenetic tree of prophages identified in D. mccartyi closed genomes including those from KB-1. Most likely tree of 100 bootstraps, scale indicates number of nucleotide substitutions per site. Spacer matches are highlighted with red bars (KBDCA3 I-C system) or blue bars (KBVC1 I-E system) with spacer number indicated below hit. Matches from all D. mccartyi spacers are provided in Table S3. Two incomplete and two highly similar D. mccartyi prophages were omitted but can be found in Table S4. Nucleotide sequences are provided as supplemental File S3. Sequences corresponding to prophage morphological proteins are abbreviated by name in first instance and subsequently color coded: ST small terminase, LT large terminase, PO portal, HP head protease, HD head decoration, MH major head, PC packaging chaperone, TC tail connector, TR tail terminator, TT tail tube, TG tail assembly chaperone, TM tail tape measure, HN HTH endonuclease, SA recombinase, DT tail protein, BH baseplate, HS head scaffold, 6F T6SS IcmF, RP replication protein, VV DSMS3 protein, IN phage integrase, CR HTH repressor, LM tail tip, PG PG hydrolase, C1 connector 1, EP encapsulin packaged protein, S4 PMR-associated domain, HC hcp1 Type VI SS