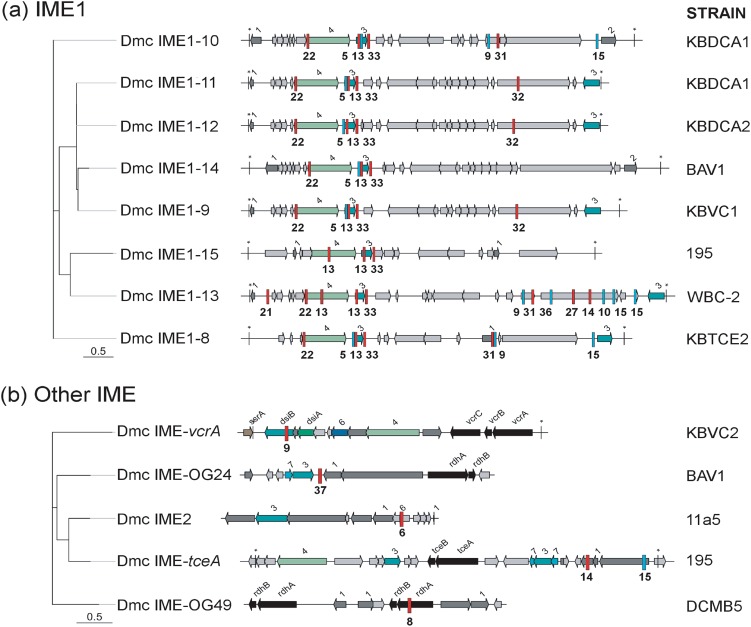
Fig. 3.
Maximum likelihood phylogenetic tree of integrative mobilizable elements (IMEs) targeted by KBVC1 and KBDCA3 CRISPR-Cas systems. KBVC1 CRISPR spacer matches as blue bars with spacer number underneath, KBDCA3 spacer matches as red bars. a D. mccartyi (Dmc) IME1 family and b other IMEs. Most likely tree of 100 bootstraps, scale indicates number of nucleotide substitutions per site. Coding regions are shown as arrows and are identified by name, or by number as follows: 1—transcriptional regulator; 2—phage antirepressor; 3—recombinase/integrase; 4—Phage DNA primase/polymerase; 6—DNA segregation protein; 7—transposase. A star (*) indicates flanking repeat region. See Table S3 for tabular version of spacer matches to targets. Gray arrows are hypothetical coding regions